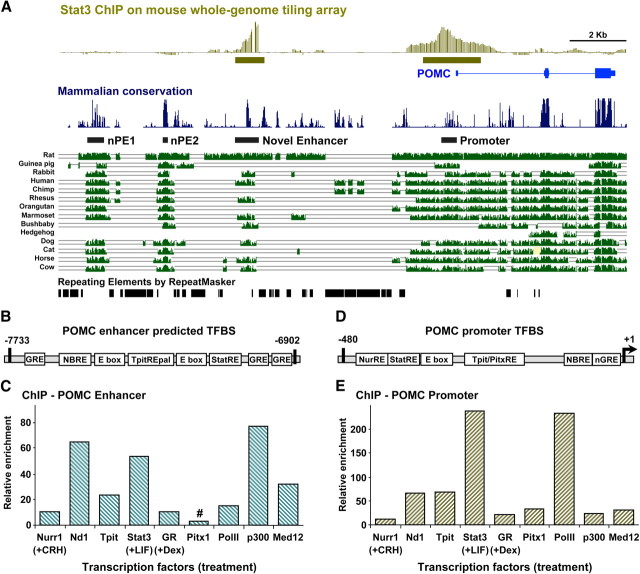
Fig. 1.
Identification of a novel POMC regulatory region. A, Whole-genome Stat3 ChIP-chip analyses in AtT-20 (18) cells reveal recruitment of Stat3 at the POMC promoter and in a region located about 7 kb upstream (green boxes, top diagram). Blue boxes and lines represent the POMC gene exons and introns, respectively. The “mammalian conservation” diagram represents sequence conservation around the POMC locus derived from 19 available mammalian species using mm9 assembly on the UCSC genome browser (32, 33). The position of previously identified hypothalamic regulatory elements nPE1 and nPE2 (16) as well as that of the promoter and −7-kb enhancer are indicated by black boxes. B, Schematic representation of the −7-kb enhancer indicating putative regulatory elements and transcription factor binding sites as identified by in silico analyses with MatInspector software (Genomatix) C, ChIP analyses of the −7-kb POMC enhancer for recruitment of the indicated transcription factors. ChIP experiments performed in AtT-20 cells used hormone-stimulated cells for Nurr1 (+CRH), Stat3 (+LIF), and GR (+Dex). ChIP enrichments are normalized relative to a control IgG ChIP and an unbound control region of the MyoD gene. Data are representative of at least three independent experiments. #Pitx1 ChIP is the only one to exhibit no significant recruitment. D, Schematic representation of the POMC promoter indicating the position of known regulatory elements. E, ChIP analyses of transcription factor recruitment to the POMC promoter in the same experiment as presented for the enhancer in C. All recruitments are significant compared with IgG control.