Figure 2.
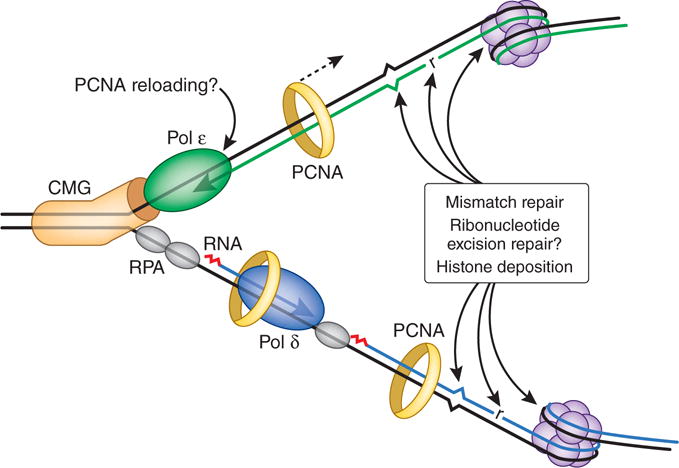
The eukaryotic replication fork. Modeled after ref. 2, but with postreplicative DNA transactions included. The CMG complex (orange) unwinds parental DNA to permit continuous leading-strand synthesis by Pol ε (green). The lagging strand is synthesized as a series of Okazaki fragments that are initiated by an RNA primer (red) made by a primase. This RNA primer is then extended by Pol α to generate a DNA primer that is extended by Pol δ (blue), which in turn synthesizes most of the nascent lagging strand with the assistance of PCNA (yellow ring). Given the weak PCNA–Pol ε interactions, we indicate the possibility of occasional dissociation of PCNA in order to mark the leading strand4 for postreplicative events that require PCNA. The most frequent of these events is histone deposition to initiate assembly of nucleosomes (purple), which are present every few hundred base pairs. Ribonucleotides (r) are incorporated during replication once every 1,000 to 10,000 bp and are removed by the ribonucleotide excision repair pathway, which could theoretically be coordinated with replication. Least abundant yet highly important are those mismatches generated by the replicases that escape proofreading and that must be removed by mismatch repair to suppress genome instability and tumorigenesis. RPA, replication protein A.
