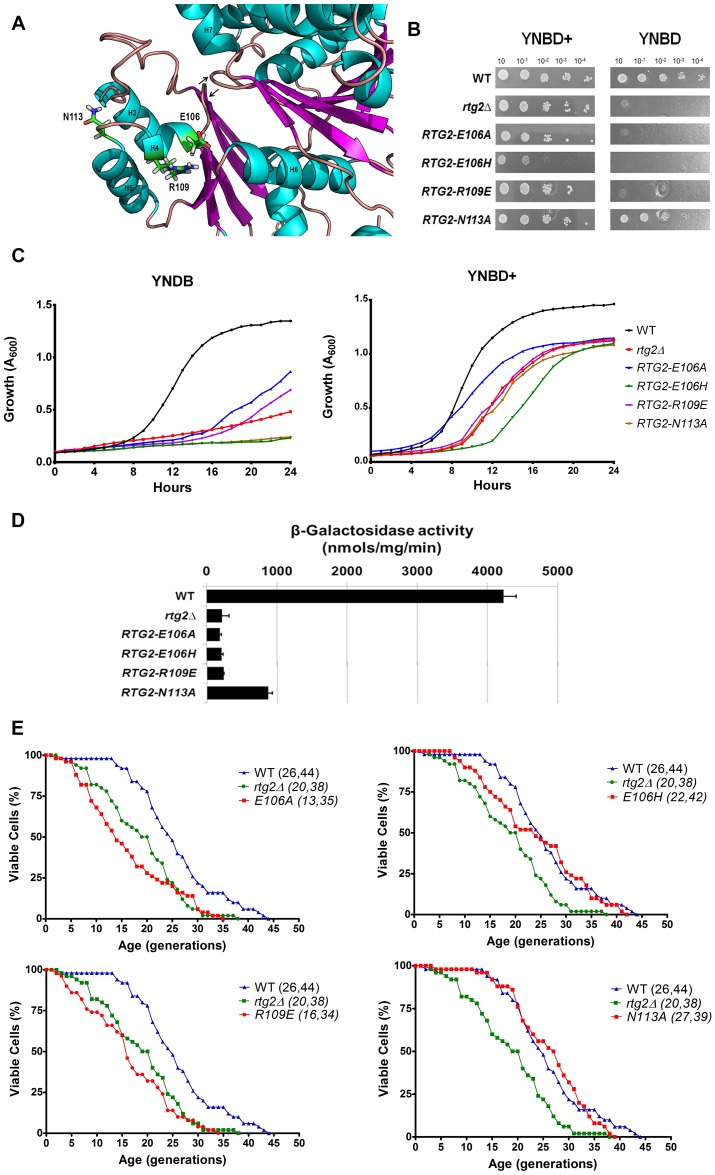
Fig 4. Mutations in Rtg2p surface residues E106, R109 and N113 impair protein function.
(A) Location of E106, R109 and N113 residues on Rtg2p protein structural model. ATP-binding loops are indicated with arrows. (B) Mutant strains RTG2-E106A, RTG2-E106H, RTG-R109E but not RTG2-N113A are auxotrophic for glutamate. Auxotrophy assays were performed on solid minimal media YNBD and YNBD+ (YNBD plus glutamate at 0.02%) by spotting serial dilutions of cells that were incubated at 30°C for three days. (C) Growth performance of strains on minimal liquid medium. Wild type, rtg2Δ and point mutants strains were grown on YPD until saturation, diluted to A600 = 0.1 in 200 μL of either YNBD or YBND+, and growth was monitored as described in Materials and Methods. (D) RTG2-N113A mutant strain retains 21% of wild type CIT2-LacZ expression in minimal medium. Wild type, rtg2Δ and point mutants were grown on YNBD medium until A600 = 0.6 and cell extracts were prepared to analyze CIT2-LacZ expression. Triplicate β-galactosidase assays were performed and the activity normalized by total protein concentration. (E) Longevity assays show that lifespan of mutant strain RTG2-E106A (top left panel) is significantly reduced, which is not observed in RTG2-E106H (top right panel), RTG-R109E (bottom left panel), and RTG2-N113A (bottom right panel) strains. Fifty cells of each strain were aligned on YPD and daughter cells were removed from mothers to construct survival curves from at least two independent experiments. The mean and maximum longevity are indicated between parentheses (mean, maximum). Statistical significance between samples is summarized in Table 2.