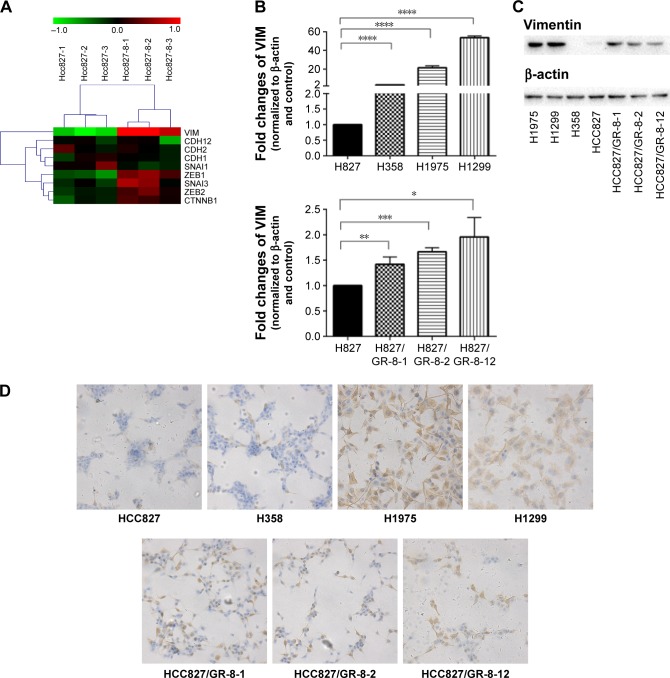
Figure 2.
Among the genes analyzed, nine of those differentially expressed are known to influence EMT.
Notes: (A) Marked upregulation of VIM mRNA (encoding vimentin, which is associated with EMT) was observed in HCC827/GR-8-1 cells. (B) Quantitative RT-PCR analysis demonstrating that the expression levels of VIM were upregulated in cells with intrinsic gefitinib resistance (H1975 and H1299) (****P<0.0001) and in those with acquired gefitinib resistance (HCC827/GR-8-1, HCC827/GR-8-2, and HCC827/GR-8-12) (*P<0.05; **P<0.01; ***P<0.001). (C) Vimentin protein was upregulated in GR cells (H1975, H1299) and in those with acquired gefitinib resistance (HCC827/GR-8-1, HCC827/GR-8-2, and HCC827/GR-8-12). (D) Immunohistochemistry demonstrated a marked increase of the mesenchymal marker, vimentin, in H1299, H1975, H358, HCC827, and HCC827/GRs cells (200×).
Abbreviations: EMT, epithelial-to-mesenchymal transition; GR, gefitinib-resistant; RT-PCR, real-time polymerase chain reaction; VIM, vimentin.