Abstract
Protein footprinting mediated by mass spectrometry has evolved over the last 30 years from proof of concept to commonplace biophysics tool, with unique capabilities for assessing structure and dynamics of purified proteins in physiological states in solution. This review outlines the history and current capabilities of two major methods of protein footprinting: reversible hydrogen-deuterium exchange (HDX) and hydroxyl radical footprinting (HRF), an irreversible covalent labeling approach. Technological advances in both approaches now permit high-resolution assessments of protein structure including secondary and tertiary structure stability mediated by backbone interactions (measured via HDX) and solvent accessibility of side chains (measured via HRF). Applications across many academic fields and in biotechnology drug development are illustrated including: detection of protein interfaces, identification of ligand/drug binding sites, and monitoring dynamics of protein conformational changes along with future prospects for advancement of protein footprinting in structural biology and biophysics research.
Introduction to Footprinting: Roots and History
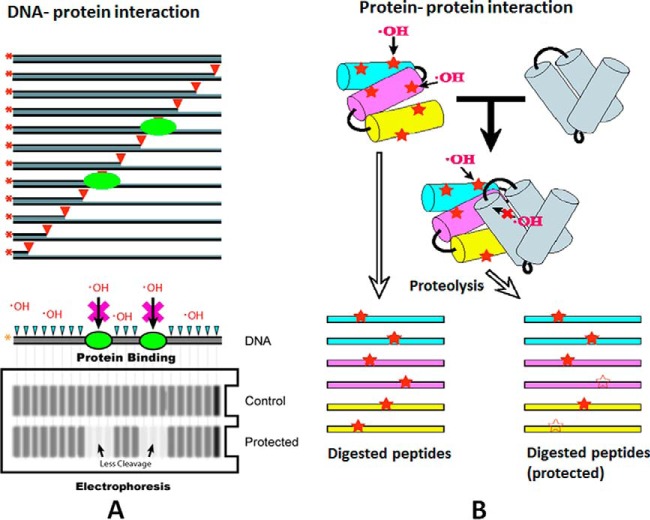
“Footprinting” based technologies were initially developed to understand nucleic acids structure and dynamics. Initially enzymatic cleavage of DNA was used to sample “exposed” sites revealing the “protected” DNA binding site for a protein-DNA complex (1) The precision and accuracy of the methods were later refined by Galas and Schmitz establishing footprinting as a quantitative biophysics tool for studying macromolecular interactions (2, 3). Over time chemical labeling approaches using small reagents, including the hydroxyl radical, (Fig. 1A) were developed to precisely define the boundary between exposed and buried regions and to extend footprinting even to in vivo studies (4–6). Early protein footprinting studies included hydrogen exchange footprinting to examine protein structure (advanced by Rosa and Richards) (7, 8) whereas alternative approaches to probe conformational change used enzymatic cleavage and gel analysis (e.g. similar to those of nucleic acids footprinting) to successfully define Ab epitopes (9).
Fig. 1.
Footprinting strategy for mapping DNA-protein binding via cleavage (left) and protein-protein interactions via modification (Right). A, Proteins (Green) interact with labeled DNA. In control experiments, all nucleotide sites can be cleaved and the DNA gel shows an uninterrupted ladder of fragments with constant intensities. In the protected lane, the position of the bound proteins are revealed by decreases in band intensities. B, Protein modification sites are observed on the pictured 3-helix bundle; these modified sites are distributed onto peptides upon protease cleavage. When a protein interacts with another protein, reduced modification occurs on the peptides (and their modification sites) that form part of the interaction surface.
Advances in mass spectrometry revolutionized protein footprinting starting in the early 1990s. For example, in chemical labeling studies of surface accessible lysine residues (10, 11) mass spectrometry methods were employed to identify modification sites based on mass shift (Fig. 1B). At the same time the hydrogen-deuterium exchange (HDX)1 field adopted mass spectrometry introducing intact protein exchange MS (12, 13) as well as localization of exchange through cleavage and peptide analysis by MS (13). As protein footprinting techniques evolved (similar to the development of nucleic acids footprinting), enhancing structure resolution and measurement accuracy were needed to move the field forward. In addition, footprinting experiments typically need to balance the need for comprehensive labeling (required to get a good signal across many sites) while avoiding artifacts of the labeling process itself. In this respect HDX-MS is advantageous as it represents a minimal perturbation of the protein. For other protein labeling methods, which are typically irreversible, a detailed understanding of the chemistry of the individual reagents, development of appropriate dosimetry measurements, and developing quantitative mass spectrometry readouts of modified species with adequate dynamic range have been critical to progress. In this review, we discuss the evolution of reagents and MS methods that now allow accurate medium to high-resolution assessments of protein structure in solution to be routinely completed using micrograms of protein samples that can be poised in a wide range of biochemical states.
Description of Labeling Technologies for Footprinting: Deuterium and OH Radical Labeling Probes Backbone and Side-chain Structure of Proteins
HDX-MS accurately reports stability of the hydrogen bonding of backbone amide residues by measuring isotopic exchange of hydrogen with its deuterium isotope (14). In continuous HDX labeling a target protein solution is labeled by dilution or rapid buffer exchange in pure D2O to produce final deuterium concentrations greater than 95% (15). Exchange is monitored for time periods ranging from seconds to days until the desired level of exchange takes place. Aliquots of the reaction are acid quenched at selected time points and analyzed by LC-MS subsequent to protease cleavage separately. Peptide mass values and intensities are extracted from the chromatogram and mass shifts are monitored versus time as changes in the centroid mass (Fig. 2A).
Fig. 2.

Flow chart of HDX-MS (Top) and HRF-MS (Bottom). A, Accessible hydrogen on backbone of the target protein are exchanged by deuterons after incubating with D2O on timescales ranging from seconds to days and the exchange is quenched by lowering pH and temperature of the reaction buffer. The uptake of deuterons is measured by use of rapid LC separation (∼mins) and mass spectrometry detection after fast pepsin digestion (∼mins) at low temperature. HDX-MS curves are plotted by deuteron uptake (as a number or %) as a function of exchange time. B, The target protein is exposed to hydroxyl radical labeling and the reaction is stopped by addition of quenchers (e.g. 10 mm methionine amide). The extent of oxidation is measured by LC separation (∼hours) and mass spectrometry after proteolysis under optimized conditions. HRF dose response curves are plotted as a fraction of unmodified peptides versus exposure time.
Hydroxyl radicals suitable for HRF labeling can be generated by various methods such as radiolysis of water with electrons, x-rays or gamma radiation, photolysis of peroxide, transition metal-dependent chemical reactions with peroxide, or high voltage electrical discharge in water (16–28). The hydroxyl radicals abstract hydrogen from aliphatic residues and directly attack sulfur atoms and aromatic rings (29). The peptide-radical species generated are efficiently quenched by oxygen in solution leading to the final products detected by MS. Typically, in HRF-MS a target protein solution is labeled for defined times and the extent of unmodified and modified species is determined after protease cleavage by mass based extraction from the chromatogram for peptides of interest (Fig. 2B). Compared with chemical reactions such as Fenton chemistry (30) where minutes of exposure to labeling reagents is required, synchrotron radiolysis (29, 31, 32) and laser photolysis approaches (33, 34) have microsecond to millisecond exposure times.
Experimental Approaches
a. Bottom-up Proteomics to Deconstruct, Extract, and Quantify Structural Information in Footprinting Experiments
Subsequent to labeling of the target protein, mass spectrometry-based bottom up proteomic approaches are applied to identify and quantitate peptides across the protein sequence that accumulate mass increases as a function of labeling. The ability to associate specific mass accumulations with specific peptides generated by protease cleavage provides details of local structure. Given the special conditions required for minimizing back-exchange in HDX-MS, acidic tolerant proteases such as pepsin are widely used to digest deuterated proteins at pH 2.5 (where exchange is minimized) in an on-line format using immobilized enzyme that provides rapid and reproducible cleavage in minutes (35, 36). Aside from rapid protease digestion, the accuracy and precision of HDX-MS data has benefited from the use of ultra-high pressure reverse phase chromatography systems (UPLC) that provide efficient and fast separation of peptide fragments (∼6 min) at reduced temperature (0 °C) (35).
Because oxidative modifications are stable and irreversible, the downstream analysis for HRF-MS, especially direct tandem MS detection of modified sites, are more flexible than for HDX. Digestion conditions can be optimized for maximal coverage (e.g. selections of denaturant, reducing reagents, cleanup procedures and enzyme combinations). After digestion, peptide mixtures containing both unmodified and modified species are separated by reverse phase LC and identified by MS. The most common oxygen adducts (+14, +16 Da) usually elute prior to unmodified species because of the increased polarity conferred by the addition of oxygen whereas Arg or His oxidized species elute later than the unmodified species owing to different chemistries involved. By comparison with HDX-MS, hydroxyl radical labeling chemistry is complex, and additional limitations include potential scavenging of radicals by solutes such a sugars, glycerol, lipids, nucleotides etc. that can interfere with labeling desired side chain targets.
b. Data Analysis and Interpretation of Mass Spectrometry-based Footprinting Data
In HDX, deuterium incorporation can be analyzed by the “centroid” or the “theoretical fit” method. The centroid method measures the weighted m/z average of each peptide distribution over the experimental time course (13). Theoretical fit method determines the number of incorporated deuterium of a HDX peptide by fitting multiple theoretical isotopic envelopes of the peptide consistent with the observed isotopic distribution using least square regression (37, 38). Fig. 3A illustrates H/D exchange data (number of deuterons as a function of exchange time) when ADAMTS13 (a disintegrin and metalloproteinase with a thrombospondin type 1 motif) is examined in the presence (red) and absence (green) of monoclonal antibodies (39). The proposed epitope includes five- loop regions (Fig. 3B) with slowed rates of exchange upon Ab-antigen binding. The y axis in Fig. 3A reflects the number of deuterons added to the peptide whereas the x axis reflects the time of exchange. Faster apparent rates of exchange reflect decreases in stability of the involved backbone residues, in this case reflected as the sum of their properties.
Fig. 3.
Typical data showing changes in labeling of a specific peptide in HDX-MS as a function of time and typical processed selected peptides comparing two conditions. A, Comparative data for the H/D exchange behavior of various peptides in the N-terminal domain of the ADAMTS13 protein in the presence (red) and absence (green) of scFv4–20, the single chain fragments of variable regions from the tested antibody sequences (39). B, Representation of binding epitopes (Red) with the reduced HDX rates in five- loop region in the spacer domain of ADAMTS13.
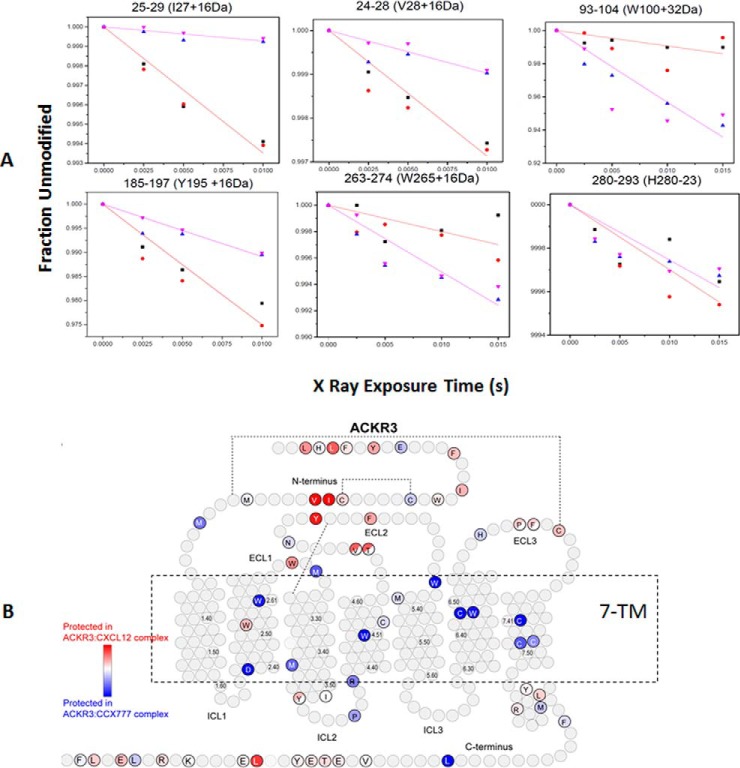
In HRF-MS peptide modification extents and rates (k) are calculated using exact mass based extracted ion currents from LC-MS. The fraction of unmodified or modified peptide is derived from ratio of chromatography peak area under the unmodified/modified species to total peak areas of all the species (including unmodified and modified). Modification rates of a peptide can be calculated from the dose-response curve plotted by exposure times (t) versus unmodified fraction of the peptide (y (t)) which follows a pseudo-first-order function: y(t) = e−kt. Fig. 4A illustrates typical data for the labeling of ACKR3 (atypical chemokine receptor 3) bound with two different ligands (chemokine CXCL12 and small molecule CCX777) as a function of exposure time (40), the y axis here reflects the degree of “unmodified” peptide whereas the x axis reflects the labeling time. It was discovered that residues in N terminus and ECL2 of ACKR3 have rates of modification reduced by CXCL12 binding whereas multiple residues in the transmembrane region vary in the case of the CCX777 complex as shown in Fig. 4B.
Fig. 4.
Typical data showing changes in peptide based oxidation rates in HRF-MS. A, Comparative data as dose response curves for oxidative labeling of ACKR3 in complex with CXCL12 or CCX777. Purple and blue triangle are replicates of ACKR3:CXCL12, red circles and black squares are replicates of ACKR3:CCX777. B, Oxidative labeling results are mapped on 2D topology plot of ACKR3. Residues are colored using a gradient where red colored residues demonstrated lower oxidation rate (more protected) in ACKR3:CXCL12 than in ACKR3:CCX777 whereas blue colored residues demonstrated higher oxidation rate (less protected) in ACKR3:CXCL12 compared with ACKR3:CCX777. Gray colored residues are not modified and half-colored circles indicate that an oxidation labeled one or both of two adjacent residues. The regions within the dashed line are the seven transmembrane domains of ACKR3.
c. Prospects for automation
Data acquisition and analysis of large-scale structural MS data in protein footprinting experiments is a cumbersome task. Promisingly, integrated software has been developed to ease the burden of data analysis. MagTran (41) is a software that calculates centroid m/z for HDX peptides. HX-Express is a semiautomated data processing HDX-MS data that exports deuterium incorporation rate curves and peak width plots (42). Three hierarchical HDX software packages include: the deuterator (43), HD Desktop (HDD) (44), and HDX Workbench (45); these provide more automated features including web-based user interfaces for entering raw data, visualization of complete HDX-MS data and results, and comparison of multiple projects. TOF2H is designed to automate data analysis of MALDI-based HDX (46). HDX analyzer (47) is developed to evaluate structural dynamics changes using HDX data with multiple statistical methods included (R, Python, and RPY2).
ProtMapMS (www.neoproteomics.net) is a software that integrates identification of peptide modifications, dose response curves plotting and oxidation rate calculation for HRF data analysis (48). Another software, ByologicTM (www.proteinmetric.com) can identify protein modifications/sequence variants and visually compares MS1 and MS2 spectra of modified and unmodified forms (49). Its capability of accurate relative quantitation of oxidized variants relative to the unmodified sequence provides utility in HRF data analysis.
Use of Structural Mass Spectrometry Data for Structural Assessment
a. Footprinting Provides Protein/Protein Interface Mapping
Binding interfaces involved in protein-protein interactions typically occlude solvent and stabilize protein secondary and tertiary structures of proteins in a complex. Thus, observed reductions in labeling rates upon protein complex formation in protein footprinting experiments (HDX-MS or HRF-MS) are used to identify potential binding sites (50). Many successful examples of mapping binding interfaces have been published to date. However, as footprinting only reports local information and cannot distinguish direct binding from allosteric effects of binding, other strategies such as mutagenesis, homology modeling, cross-linking or other structural methods are combined to support an understanding of the exact nature of the binding epitopes. For example, a combination of HDX-MS and mutagenesis was used to identify the binding interfaces between auto-antibodies derived from patients with acquired thrombotic thrombocytopenic purpura and ADAMTS13, the host metalloproteinase target (39). Epitopes in five-loop regions with slowed HDX rates upon binding were detected by HDX MS (Fig. 3B) whereas mutations in the five proposed binding loops of ADAMTS13 eliminated antibody binding and provide support for the proposed epitope. HDX-MS, like X-ray crystallography, can also identify complex discontinuous epitopes that are missed by linear methods such as peptide scanning and phage display. This is clearly illustrated in a study that defined the binding of factor H binding protein (fHbp), a key virulence factor and vaccine antigen of Neisseria meningitides, with its bactericidal mAb 12C1 (51) as the MS and crystallography data delineated roles for both N-terminal and C-terminal peptide regions. Both HDX-MS and HRF-MS were used to map the epitope of human epidermal growth factor receptor (EPGR) that binds to adnectin, a targeted biologic derived from the fibronectin domain, these solution data were consistent with that from crystallography (52, 53).
Protein footprinting techniques to probe protein-protein interactions have been used heavily in biotechnology research, these studies are quite important for verifying epitopes and paratopes in monoclonal antibody drug development (54). In addition, protein footprinting is valuable for comparing structures of “innovator” therapeutic proteins and their biosimilars; this can help establish the potential equivalency of the biosimilars with the patented drugs. For example, HDX-MS was used to characterize the structure of various versions of insulin (55, 56) and several forms of glucocerebrosidase (brand name Cerezyme) for treatment of Gaucher disease (57). Likewise, HRF-MS was used to analyze conformations of therapeutic proteins including recombinant erythropoietin (EPO), interferon α-2A (IFN), granulocyte colony-stimulating factor (GCSF) (58).
b. Footprinting Provides Protein-Drug Interaction Information
Small molecule binding to proteins does not bury a large surface area as in protein-protein interactions, however protein footprinting can reveal these drug-occupied surface or near surface pockets. For example, HDX-MS was used to analyze small molecule binding to a functioning PPARγ (peroxisome proliferator-activated receptor gamma) (59). HDX analysis of one such synthetic compound (SR1664) revealed that in contrary to agonists, SR1664 binding increased the mobility of the PPARγ C-terminal end helix (H11), which is close to another helix (H12) contributing to hydrogen bonds of PPARγ and full agonists (60). In the glioma field the drug-binding region of a potent inhibitor (SCB4380) targeting PTPRZ (protein tyrosine phosphatase receptor-type Z) was identified by HDX-MS in catalytic pocket of PTPRZ-D1 (61). Likewise, HRF-MS analysis of serotonin type 4 receptor (5-HT4R) in the presence and absence of an antagonist revealed the ligand-binding pocket consistent with data from other GPCRs and was used as well to define mechanisms of signal transmission (62). Also using HRF-MS, a small molecule partial agonist of atypical chemokine receptor 3 (ACKR3) was mapped to its binding site (Fig. 4B) along with the details of the receptor's binding of chemokine CXCR12 (Fig. 5A) (63). Overall, protein footprinting is seen to be very valuable in mapping the binding of both small and large molecules to proteins for both academic research and drug development.
Fig. 5.
An example of the use of HRF-MS to probe absolute structure changes of receptor-chemokine interactions. A, Mapping of protein footprinting oxidation labeling results on topology plot of CXCL12. Red colored residues indicate reduced oxidation rates in the ACKR3:CXCL12 complex versus CXCL12 alone, gray colored residues are not modified and half-colored circles indicate a modification on one or both of two adjacent residues. B, Natural log of protection factors derived from experimental protein footprinting data on CXCL12 alone showed linear correlation with the fractional side chain SASA (0 corresponds to no solvent exposure and 1 corresponds to complete exposure) calculated from the crystal structure of CXCL12 (PDB: 1A15). C, Mapping of calculated SASA values (from regression plot) for CXCL12 free state on the crystal structure. D, Mapping of predicted SASA values for CXCL12 in complex with ACKR3 based on interpolation from the linear correlation in B on the chemokine crystal structure (63).
c. Footprinting is a Structural Kinetic Probe of Protein Conformational Changes
Kinetics investigations of protein folding and dynamics are important applications of MS-based protein footprinting. The protein folding of α1-antitrypsin was studied by pulse-labeling HDX-MS, where the protein unfolding and refolding was performed by addition and dilution of denaturant (64). Deuteration of the unfolded part of the protein measured at each folding time was interpreted in the context of existing crystal structures. A study of chaperone (GroEl/ES)-assisted protein (TIM-barrel) folding by use of pulse-labeling HDX-MS showed acceleration of the protein folding by the chaperones (65). HDX-MS has also been used for studies of protein aggregation or fibril formation resulting from improper protein folding (66) and pulse-labeling HDX-MS was used to monitor refolding of bacteriorhodopsin induced by the denaturant (67). Quench flow HDX-MS can be used access millisecond time scale dynamics as seen in cytochrome-c refolding and other protein dynamics studies (68, 69).
HRF-MS can monitor protein conformational changes from microseconds to minutes. A recent radiolysis-based HRF-MS study of YiiP, a proton-coupled Zinc (II) transporter, elucidated the millisecond conformational changes of water accessibility in the TM5 domain during Zinc (II) binding (70). Time-resolved synchrotron radiolytic HRF was also employed to study the dynamic conformational changes in gelsolin upon Ca (II) binding (71) on the millisecond timescale. In both these studies, introduction of metal ions to apo-protein via rapid mixing was initiated, and delay times from a few milliseconds to a few minutes (after mixing) were introduced prior to exposure of samples by radiolysis.
A combination of HDX-MS, HRF-MS and another structural technique, small angle X-ray scattering (SAXS), was employed to investigate the conformational changes of orange carotenoid protein (OCP) upon photo-activation (72). In addition to global structural changes revealed by SAXS, local conformational changes providing a detailed model of the conformational dynamics were detected by HDX-MS and HRF-MS. Detection of decrease in solvent accessibility of carontenoid binding residues in N-terminal domain and increases in C-terminal domain, together with crystal structural data of the OCP and RCP (red carotenoid protein) revealed a 12 angstrom translocation of carotenoid pigment within the protein upon OCP photoactivation including time resolved analysis of the reaction (73). This work is an excellent example of the merging of local detailed structural information on dynamics provided by footprinting with global crystallographic and SAXS signatures.
Stocks et al. (74) combined stopped-flow mixing and pH jump to observe unfolding of cytochrome-c, holo-myoglobin, and S100A11 in the 10 ms range; HRF-MS was achieved by laser photolysis of peroxide at time points of interest after mixing. A laser induced temperature jump coupled with laser photolysis of peroxide (called FPOP or fast footprinting of proteins) was implemented by Gross et al. in probing folding dynamics of barstar on the sub-millisecond time scale (75). Employing sub-millisecond laminar flow mixing coupled with HRF-MS, investigation of apomyoglobin folding revealed that sub-millisecond structure formation is primarily driven by hydrophobic side chain interaction not backbone hydrogen bonding (76). Additionally, FPOP HRF-MS was used to differentiate conformational difference of Im7 (native, partially folded and globally unfolded) providing information of individual side chains in folding process (77).
New Opportunities
a. High Resolution Structure Assessment
Achieving the maximum possible structural resolution for protein footprinting approaches has been an important goal of the field, and has now been achieved by combinations of several strategies. We introduce a figure of merit for comparing structural resolution across protein footprinting experiments defined as the number of probe sites identified and quantified divided by the total number of residues, expressed as a %. This analytic view is important as protein footprinting may achieve 100% “coverage,” in that peptides that cover the entire protein sequence may be detected and analyzed, but sub-peptide level readouts may not be available to probe the fine details of the molecular interactions unless special care is taken. We examined over 60 manuscripts reporting 75 footprinting results primarily from the last 10 years to understand the evolving approaches across many laboratories. supplemental Table S1 summarizes the survey results, including 38 HDX-MS results and 31 HRF-MS results (16 using laser photolysis of peroxide and 15 using radiolysis), and seven other covalent labeling-MS results (1 acetylation, 3 GEE and 3 carbene labeling). Overall, using the above figure of merit, HRF-MS achieved structural resolution of 40% or more in many cases, whereas HDX-MS in many cases attained 100% structural resolution. These levels of structural resolution have been achieved in part through improvements in LC instrumentation (particularly UPLC) to separate peptides more efficiently and more quickly. For HRF-MS use of multiple proteases including pepsin has been effective in enhancing structural resolution whereas for HDX-MS, the development of approaches that generate overlapping peptic fragments has been quite successful (78). All these methods quantify the peptide intensities at the MS1 level. The “holy grail” of single residue structural resolution conceptually involves quantification at the MS2 level, through peptide fragmentation and tandem ion quantification. Significant progress along these lines has also been achieved by both HRF-MS and HDX-MS.
Reaching single residue structural resolution via MS2 has been challenging for HDX-MS owing to hydrogen scrambling during CID fragmentation. However, in certain cases good structural resolution was achieved by with minimal scrambling via ETD fragmentation for β2-microglobin (structural resolution = 43%) (79) and staphylococcal nuclease (structural resolution = 66%) (80). In both cases, careful detuning was used to avoid scrambling during isolation, which reduced the sensitivity of the approach. Also, charge bias (> = 3+) for peptides that are easily detected in ETD/ECD can result in incomplete sequence coverage for some large proteins, limiting application of the method.
In HRF-MS application of UPLC methods has resolved isobaric peptides where the oxidative modification exists on differing residues. In these cases, a single type of modified species, eluting at a defined retention time and with a characteristic mass, can be extracted to provide single residue footprinting data. These UPLC approaches, in combination with pepsin digestion and denaturation, were applied in HRF-MS footprinting analysis of β-amyloid via radiolysis achieving 35% structural resolution (81). Using laser photolysis based HRF-MS, investigators have achieved 40% structural resolution for thrombin (82). MS2 approaches are more feasible for HRF-MS, as scrambling is minimal, such quantification approaches have achieved 43% structural resolution for calmodulin (83). Quantitation of the labeling in this case was performed by combined analysis of both MS1 and MS2 intensities of target peptides as modification induced fragmentation effects can interfere with the analysis. Footprinting structural resolution can also be extended by use of alternative labeling reagents, such as GEE, which labels D and E residues (84, 85); these can add up to 10% to the structural resolution depending on the target sequence. Carbene mediated protein footprinting has also been reported as an alternative to label residues that are less reactive to hydroxyl radicals such as Asp, Glu (86). and carbene labeling designed on a FPOP platform resolved different structural domains of apo and holo calmodulin (86). A synthesized bespoke diazirine -based carbene footprinting with high labeling efficiency were developed to probe the substrate-binding sites of lysozyme and deubiquitinating protease (87).
b. Measuring Quantitative Structural Parameters with Footprinting
Typical footprinting measurements provide a relative comparison between two states of interest. However, inferring absolute structure parameters is possible using information from covalent labeling. For example, measured HRF-MS oxidation rates can be converted to protection factors (PF) using the known intrinsic reactivity of amino acid side chains for hydroxyl radicals (88). These HRF-based PFs show statistically significant correlations with structural factors such as solvent accessible surface area and number of local structural contacts. This was illustrated at the single-residue level for Ca2+-calmodulin, where the solvent accessible surface area values side chains was predicted from footprinting data (83). Fig. 5 shows an example of applying this new approach, where the crystal structure data of a chemokine (CXCL12) was compared with the natural log PF values derived from experimental HRF MS data (Fig. 5A, 5B, and 5C). The resulting correlation provided a molecular ruler for inferring the absolute extent of protection of the chemokine when bound to its receptor (ACKR3) as shown in Fig. 5D (63). This combination of high structural resolution and quantitative structure assessment places HRF-MS in a unique position to assess structure in absolute versus relative terms, the previous default approach for footprinting.
c. Footprinting in Cells
A longstanding goal of footprinting studies is the examination of macromolecular structure inside living cells. Although this may be out of reach using exchangeable reagents such as deuterium, it is a realistic prospect in the case of irreversible labeling. For nucleic acids footprinting, dimethylsulfate was developed as a footprinting reagent for detection of protein-DNA interactions in cells (89, 90); these approaches have been considerable improved over time (91, 92). Radiolytic footprinting has recently been used to monitor ribosome assembly in bacteria through monitoring of RNA based protections as a function of assembly (93–95) providing novel and powerful readouts of complex cellular structures.
Mass spectrometry to examine protein interactions in cells is appropriately dominated by progress in the cellular cross-linking field (96–99); however, such studies do not provide the level of molecular detail presumed for footprinting, where high coverage of the molecule is the goal. For protein footprinting in cells, the challenges are quite substantial and include identification of reagents that can penetrate cells and modify protein species efficiently coupled to recovery of sufficient material for MS detection of multiple labeled sites per protein. Hydroxyl radicals are a useful reagent for in vivo footprinting, but the low ambient oxygen levels inside cells reduces oxidation efficacy relative to in vitro studies. Possible approaches to overcome these limitations have been explored by Espino et al., who used laser photolysis of peroxide to oxidize proteins inside cells (100). In this case the peroxide was able to efficiently penetrate cells and native enzymes, like superoxide dismutase, provided a source of oxygen. Future approaches will certainly require multidimensional separations, targeted proteomic analysis and pull-down strategies to enhance coverage and provide functional information on cellular structures of interest.
CONCLUSION
Protein footprinting has evolved considerably over 30 years and its growth has occurred hand in hand with the significant advances in mass spectrometry that have occurred during the same time. At present, protein footprinting is making unique contributions to understanding protein structure and dynamics for macromolecular complexes, membrane proteins, and bio-therapeutics: three of the most challenging and important areas of research in structural biology today. In particular, the modest sample requirements and flexibility in use of solution conditions for conducting footprinting analysis are two of its greatest advantages. Thus, difficult studies such as high-resolution structural kinetic analysis of protein dynamics or examination of the structure of native membrane protein forms are feasible by these methods. Recent advances in technique have pushed the structural resolution closer to the maximum possible, but further increases in signal to noise for detecting labeled species are needed to address more complex systems routinely.
Supplementary Material
Acknowledgments
We thank Dr. Janna Kiselar for scientific discussions and suggestions.
Footnotes
Author contributions: M.R.C. designed research; L.W. performed research; L.W. analyzed data; L.W. wrote the paper; M.R.C. reviewed and edited manuscript.
* This work is supported by NIH grant P30EB009998 from the National Health Institute and by the National Science Foundation grant, DBI-1228549. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental material.
This article contains supplemental material.
Conflict of interest statement: Dr. Chance is a founder and shareholder of the company NeoProteomics which has licensed technology from Case Western Reserve University in the field of mass spectrometry including the software ProtMapMS. Neo Proteomics also is a contract research organization performing commercial protein footprinting services.
1 The abbreviations used are:
- HDX
- hydrogen deuterium exchange
- Ab
- Antibody
- ACKR3
- atypical chemokine receptor 3
- HRF
- hydroxyl radical (.OH) based protein footprinting
- UPLC
- ultra-high pressure reverse phase chromatography systems
- ETD
- electron transfer dissociation
- ECD
- electron capture dissociation
- FPOP
- fast photochemical oxidation of proteins.
REFERENCES
- 1. Mirzabekov A. D., and Melnikova A. F. (1974) Localization of chromatin proteins within DNA grooves by methylation of chromatin with dimethyl sulphate. Mol. Biol. Rep. 1, 379–384 [DOI] [PubMed] [Google Scholar]
- 2. Galas D. J., and Schmitz A. (1978) DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 5, 3157–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schmitz A., and Galas D. J. (1979) The interaction of RNA polymerase and lac repressor with the lac control region. Nucleic Acids Res. 6, 111–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adilakshmi T., Bellur D. L., and Woodson S. A. (2008) Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature 455, 1268–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hulscher R. M., Bohon J., Rappe M. C., Gupta S., D'Mello R., Sullivan M., Ralston C. Y., Chance M. R., and Woodson S. A. (2016) Probing the structure of ribosome assembly intermediates in vivo using DMS and hydroxyl radical footprinting. Methods 103, 49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brenowitz M., Chance M. R., Dhavan G., and Takamoto K. (2002) Probing the structural dynamics of nucleic acids by quantitative time-resolved and equilibrium hydroxyl radical “footprinting”. Curr. Opin. Struct. Biol. 12, 648–653 [DOI] [PubMed] [Google Scholar]
- 7. Hvidt A., and Linderstrom-Lang K. (1954) Exchange of hydrogen atoms in insulin with deuterium atoms in aqueous solutions. Biochim. Biophys. Acta 14, 574–575 [DOI] [PubMed] [Google Scholar]
- 8. Rosa J. J., and Richards F. M. (1979) An experimental procedure for increasing the structural resolution of chemical hydrogen-exchange measurements on proteins: application to ribonuclease S peptide. J. Mol. Biol. 133, 399–416 [DOI] [PubMed] [Google Scholar]
- 9. Sheshberadaran H., and Payne L. G. (1988) Protein antigen-monoclonal antibody contact sites investigated by limited proteolysis of monoclonal antibody-bound antigen: protein “footprinting”. Proc. Natl. Acad. Sci. U.S.A. 85, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hanai R., and Wang J. C. (1994) Protein footprinting by the combined use of reversible and irreversible lysine modifications. Proc. Natl. Acad. Sci. U.S.A. 91, 11904–11908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Steiner R. F., Albaugh S., Fenselau C., Murphy C., and Vestling M. (1991) A mass spectrometry method for mapping the interface topography of interacting proteins, illustrated by the melittin-calmodulin system. Anal. Biochem. 196, 120–125 [DOI] [PubMed] [Google Scholar]
- 12. Katta V., and Chait B. T. (1991) Conformational changes in proteins probed by hydrogen-exchange electrospray-ionization mass spectrometry. Rapid Commun. Mass Spectrom. 5, 214–217 [DOI] [PubMed] [Google Scholar]
- 13. Zhang Z., and Smith D. L. (1993) Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 2, 522–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wales T. E., and Engen J. R. (2006) Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 25, 158–170 [DOI] [PubMed] [Google Scholar]
- 15. Engen J. R., and Smith D. L. (2000) Investigating the higher order structure of proteins. Hydrogen exchange, proteolytic fragmentation, and mass spectrometry. Methods Mol. Biol. 146, 95–112 [DOI] [PubMed] [Google Scholar]
- 16. Xu G., and Chance M. R. (2005) Radiolytic modification and reactivity of amino acid residues serving as structural probes for protein footprinting. Anal. Chem. 77, 4549–4555 [DOI] [PubMed] [Google Scholar]
- 17. Gau B. C., Sharp J. S., Rempel D. L., and Gross M. L. (2009) Fast photochemical oxidation of protein footprints faster than protein unfolding. Anal. Chem. 81, 6563–6571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Asmus K. D. (1984) Pulse radiolysis methodology. Methods Enzymol. 105, 167–178 [DOI] [PubMed] [Google Scholar]
- 19. Farhataziz and Rodgers M. A. J. (1987) Radiation Chemistry: Principles and Applications [Google Scholar]
- 20. Ottinger L. M., and Tullius T. D. (2000) High-Resolution in Vivo Footprinting of a Protein-DNA Complex Using γ-Radiation. J. Am. Chem. Soc. 122, 5901–5902 [Google Scholar]
- 21. Franchet-Beuzit J., Spotheim-Maurizot M., Sabattier R., Blazy-Baudras B., and Charlier M. (1993) Radiolytic footprinting. Beta rays, gamma photons, and fast neutrons probe DNA-protein interactions. Biochemistry 32, 2104–2110 [DOI] [PubMed] [Google Scholar]
- 22. Volman D. H., and Chen J. C. (1959) The photochemical decomposition of hydrogen peroxide in aqueous solutions of allyl alcohol at 2537 Å.1. J. Am. Chem. Soc. 81, 4141–4144 [Google Scholar]
- 23. Fenton H. J. H. (1894) LXXIII.-Oxidation of tartaric acid in presence of iron. J. Am. Chem. Soc. 65, 899–910 [Google Scholar]
- 24. Maleknia S. D., Chance M. R., and Downard K. M. (1999) Electrospray-assisted modification of proteins: a radical probe of protein structure. Rapid Commun. Mass Spectrom. 13, 2352–2358 [DOI] [PubMed] [Google Scholar]
- 25. Takamoto K., and Chance M. R. (2006) Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annu. Rev. Biophys. Biomol. Struct. 35, 251–276 [DOI] [PubMed] [Google Scholar]
- 26. Chance M. R. (2001) Unfolding of apomyoglobin examined by synchrotron footprinting. Biochem. Biophys. Res. Commun. 287, 614–621 [DOI] [PubMed] [Google Scholar]
- 27. Macgregor R. B., Jr. (1992) Photogeneration of hydroxyl radicals for footprinting. Anal. Biochem. 204, 324–327 [DOI] [PubMed] [Google Scholar]
- 28. Wang L., and Chance M. R. (2011) Structural mass spectrometry of proteins using hydroxyl radical based protein footprinting. Anal. Chem. 83, 7234–7241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu G., and Chance M. R. (2007) Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem. Rev. 107, 3514–3543 [DOI] [PubMed] [Google Scholar]
- 30. Tullius T. D., and Dombroski B. A. (1986) Hydroxyl radical “footprinting”: high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc. Natl. Acad. Sci. U.S.A. 83, 5469–5473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chance M. R., Sclavi B., Woodson S. A., and Brenowitz M. (1997) Examining the conformational dynamics of macromolecules with time-resolved synchrotron X-ray 'footprinting'. Structure 5, 865–869 [DOI] [PubMed] [Google Scholar]
- 32. Gupta S., Sullivan M., Toomey J., Kiselar J., and Chance M. R. (2007) The Beamline X28C of the Center for Synchrotron Biosciences: a national resource for biomolecular structure and dynamics experiments using synchrotron footprinting. J. Synchrotron Rad. 14, 233–243 [DOI] [PubMed] [Google Scholar]
- 33. Hambly D., and Gross M. (2007) Laser flash photochemical oxidation to locate heme binding and conformational changes in myoglobin. Int. J. Mass Spectrometry 259, 124–129 [Google Scholar]
- 34. Hambly D. M., and Gross M. L. (2005) Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J. Am. Soc. Mass Spectrom. 16, 2057–2063 [DOI] [PubMed] [Google Scholar]
- 35. Ahn J., Jung M. C., Wyndham K., Yu Y. Q., and Engen J. R. (2012) Pepsin immobilized on high-strength hybrid particles for continuous flow online digestion at 10,000 psi. Anal. Chem. 84, 7256–7262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang L., Pan H., and Smith D. L. (2002) Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol. Cell. Proteomics 1, 132–138 [DOI] [PubMed] [Google Scholar]
- 37. Hotchko M., Anand G. S., Komives E. A., and Ten Eyck L. F. (2006) Automated extraction of backbone deuteration levels from amide H/2H mass spectrometry experiments. Protein Sci. 15, 583–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Palmblad M., Buijs J., and Hakansson P. (2001) Automatic analysis of hydrogen/deuterium exchange mass spectra of peptides and proteins using calculations of isotopic distributions. J. Am. Soc. Mass Spectrom. 12, 1153–1162 [DOI] [PubMed] [Google Scholar]
- 39. Casina V. C., Hu W., Mao J. H., Lu R. N., Hanby H. A., Pickens B., Kan Z. Y., Lim W. K., Mayne L., Ostertag E. M., Kacir S., Siegel D. L., Englander S. W., and Zheng X. L. (2015) High-resolution epitope mapping by HX MS reveals the pathogenic mechanism and a possible therapy for autoimmune TTP syndrome. Proc. Natl. Acad. Sci. U.S.A. 112, 9620–9625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kiselar J. G., Janmey P. A., Almo S. C., and Chance M. R. (2003) Structural analysis of gelsolin using synchrotron protein footprinting. Mol. Cell. Proteomics 2, 1120–1132 [DOI] [PubMed] [Google Scholar]
- 41. Zhang Z., and Marshall A. G. (1998) A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass Spectrom. 9, 225–233 [DOI] [PubMed] [Google Scholar]
- 42. Weis D. D., Engen J. R., and Kass I. J. (2006) Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J. Am. Soc. Mass. Spectrom. 17 [DOI] [PubMed] [Google Scholar]
- 43. Pascal B. D., Chalmers M. J., Busby S. A., Mader C. C., Southern M. R., Tsinoremas N. F., and Griffin P. R. (2007) The Deuterator: software for the determination of backbone amide deuterium levels from H/D exchange MS data. BMC Bioinformatics 8, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pascal B. D., Chalmers M. J., Busby S. A., and Griffin P. R. (2009) HD desktop: an integrated platform for the analysis and visualization of H/D exchange data. J. Am. Soc. Mass Spectrom. 20, 601–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pascal B. D., Willis S., Lauer J. L., Landgraf R. R., West G. M., Marciano D., Novick S., Goswami D., Chalmers M. J., and Griffin P. R. (2012) HDX workbench: software for the analysis of H/D exchange MS data. J. Am. Soc. Mass Spectrom. 23, 1512–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nikamanon P., Pun E., Chou W., Koter M. D., and Gershon P. D. (2008) “TOF2H”: a precision toolbox for rapid, high density/high coverage hydrogen-deuterium exchange mass spectrometry via an LC-MALDI approach, covering the data pipeline from spectral acquisition to HDX rate analysis. BMC Bioinformatics 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu S., Liu L., Uzuner U., Zhou X., Gu M., Shi W., Zhang Y., Dai S. Y., and Yuan J. S. (2011) HDX-Analyzer: a novel package for statistical analysis of protein structure dynamics. BMC Bioinformatics 12, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kaur P., Kiselar J. G., and Chance M. R. (2009) Integrated Algorithms for High Throughput Examination of Covalently Labeled Biomolecules by Structural Mass Spectrometry. Anal. Chem. 81, 8141–8149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yong K., Marshall W. B., Eric C., Chris B., David M., and Beatrix U. (2014) Accurate and rapid quantification of co-eluting deamidated and non-deamidated peptides using a novel deconvolution technique. 64th ASMS [Google Scholar]
- 50. Mandell J. G., Falick A. M., and Komives E. A. (1998) Identification of protein-protein interfaces by decreased amide proton solvent accessibility. Proc. Natl. Acad. Sci. U.S.A. 95, 14705–14710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Malito E., Faleri A., Lo Surdo P., Veggi D., Maruggi G., Grassi E., Cartocci E., Bertoldi I., Genovese A., Santini L., Romagnoli G., Borgogni E., Brier S., Lo Passo C., Domina M., Castellino F., Felici F., van der Veen S., Johnson S., Lea S. M., Tang C. M., Pizza M., Savino S., Norais N., Rappuoli R., Bottomley M. J., and Masignani V. (2013) Defining a protective epitope on factor H binding protein, a key meningococcal virulence factor and vaccine antigen. Proc. Natl. Acad. Sci. U.S.A. 110, 3304–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Iacob R. E., Chen G., Ahn J., Houel S., Wei H., Mo J., Tao L., Cohen D., Xie D., Lin Z., Morin P. E., Doyle M. L., Tymiak A. A., and Engen J. R. (2014) The influence of adnectin binding on the extracellular domain of epidermal growth factor receptor. J. Am. Soc. Mass Spectrom. 25, 2093–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yan Y., Chen G., Wei H., Huang R. Y., Mo J., Rempel D. L., Tymiak A. A., and Gross M. L. (2014) Fast photochemical oxidation of proteins (FPOP) maps the epitope of EGFR binding to adnectin. J. Am. Soc. Mass Spectrom. 25, 2084–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Deperalta G., Alvarez M., Bechtel C., Dong K., McDonald R., and Ling V. (2013) Structural analysis of a therapeutic monoclonal antibody dimer by hydroxyl radical footprinting. MAbs 5, 86–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nakazawa S., Hashii N., Harazono A., and Kawasaki N. (2012) Analysis of oligomeric stability of insulin analogs using hydrogen/deuterium exchange mass spectrometry. Anal. Biochem. 420, 61–67 [DOI] [PubMed] [Google Scholar]
- 56. Nakazawa S., Ahn J., Hashii N., Hirose K., and Kawasaki N. (2013) Analysis of the local dynamics of human insulin and a rapid-acting insulin analog by hydrogen/deuterium exchange mass spectrometry. Biochim. Biophys. Acta 1834, 1210–1214 [DOI] [PubMed] [Google Scholar]
- 57. Tang L., Coales S. J., Morrow J. A., Edmunds T., and Hamuro Y. (2012) Characterization of the N370S mutant of glucocerebrosidase by hydrogen/deuterium exchange mass spectrometry. Chembiochem 13, 2243–2250 [DOI] [PubMed] [Google Scholar]
- 58. Watson C., and Sharp J. S. (2012) Conformational analysis of therapeutic proteins by hydroxyl radical protein footprinting. AAPS J. 14, 206–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Choi J. H., Banks A. S., Kamenecka T. M., Busby S. A., Chalmers M. J., Kumar N., Kuruvilla D. S., Shin Y., He Y., Bruning J. B., Marciano D. P., Cameron M. D., Laznik D., Jurczak M. J., Schurer S. C., Vidovic D., Shulman G. I., Spiegelman B. M., and Griffin P. R. (2011) Antidiabetic actions of a non-agonist PPARgamma ligand blocking Cdk5-mediated phosphorylation. Nature 477, 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hughes T. S., Giri P. K., de Vera I. M., Marciano D. P., Kuruvilla D. S., Shin Y., Blayo A. L., Kamenecka T. M., Burris T. P., Griffin P. R., and Kojetin D. J. (2014) An alternate binding site for PPARgamma ligands. Nat. Commun. 5, 3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fujikawa A., Nagahira A., Sugawara H., Ishii K., Imajo S., Matsumoto M., Kuboyama K., Suzuki R., Tanga N., Noda M., Uchiyama S., Tomoo T., Ogata A., Masumura M., and Noda M. (2016) Small-molecule inhibition of PTPRZ reduces tumor growth in a rat model of glioblastoma. Sci. Rep. 6, 20473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Padayatti P. S., Wang L., Gupta S., Orban T., Sun W., Salom D., Jordan S. R., Palczewski K., and Chance M. R. (2013) A hybrid structural approach to analyze ligand binding by the serotonin type 4 receptor (5-HT4). Mol. Cell. Proteomics 12, 1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gustavsson M., Wang L., Gils N. V., Stephens B. S., Mocentryx C., Yang S., Abagyan R., Chance M. R., Kufareva I., and Handel T. M. (2017) Structural basis of ligand interaction with atypical chemokine receptor 3. Nature Commnun. 8, 14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tsutsui Y., Dela Cruz R., and Wintrode P. L. (2012) Folding mechanism of the metastable serpin alpha1-antitrypsin. Proc. Natl. Acad. Sci. U.S.A. 109, 4467–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Georgescauld F., Popova K., Gupta A. J., Bracher A., Engen J. R., Hayer-Hartl M., and Hartl F. U. (2014) GroEL/ES chaperonin modulates the mechanism and accelerates the rate of TIM-barrel domain folding. Cell 157, 922–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hodkinson J. P., Radford S. E., and Ashcroft A. E. (2012) The role of conformational flexibility in beta2-microglobulin amyloid fibril formation at neutral pH. Rapid Commun. Mass Spectrom. 26, 1783–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mehmood S., Domene C., Forest E., and Jault J. M. (2012) Dynamics of a bacterial multidrug ABC transporter in the inward- and outward-facing conformations. Proc. Natl. Acad. Sci. U.S.A. 109, 10832–10836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yang H., and Smith D. L. (1997) Kinetics of cytochrome c folding examined by hydrogen exchange and mass spectrometry. Biochemistry 36, 14992–14999 [DOI] [PubMed] [Google Scholar]
- 69. Truhlar S. M. E., Croy C. H., Torpey J. W., Koeppe J. R., and Komives E. A. (2006) Solvent accessibility of protein surfaces by amide H/2H exchange MALDI-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 17, 1490–1497 [DOI] [PubMed] [Google Scholar]
- 70. Gupta S., Chai J., Cheng J., D'Mello R., Chance M. R., and Fu D. (2014) Visualizing the kinetic power stroke that drives proton-coupled zinc(II) transport. Nature 512, 101–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kiselar J. G., and Chance M. R. (2010) Future directions of structural mass spectrometry using hydroxyl radical footprinting. J. Mass Spectrom. 45, 1373–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gupta S., Guttman M., Leverenz R. L., Zhumadilova K., Pawlowski E. G., Petzold C. J., Lee K. K., Ralston C. Y., and Kerfeld C. A. (2015) Local and global structural drivers for the photoactivation of the orange carotenoid protein. Proc. Natl. Acad. Sci. U.S.A. 112, E5567–E5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Leverenz R. L., Sutter M., Wilson A., Gupta S., Thurotte A., Bourcier de Carbon C., Petzold C. J., Ralston C., Perreau F., Kirilovsky D., and Kerfeld C. A. (2015) PHOTOSYNTHESIS. A 12 A carotenoid translocation in a photoswitch associated with cyanobacterial photoprotection. Science 348, 1463–1466 [DOI] [PubMed] [Google Scholar]
- 74. Stocks B. B., and Konermann L. (2009) Structural characterization of short-lived protein unfolding intermediates by laser-induced oxidative labeling and mass spectrometry. Anal. Chem. 81, 20–27 [DOI] [PubMed] [Google Scholar]
- 75. Chen J., Rempel D. L., Gau B. C., and Gross M. L. (2012) Fast photochemical oxidation of proteins and mass spectrometry follow submillisecond protein folding at the amino-acid level. J. Am. Chem. Soc. 134, 18724–18731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vahidi S., Stocks B. B., Liaghati-Mobarhan Y., and Konermann L. (2013) Submillisecond protein folding events monitored by rapid mixing and mass spectrometry-based oxidative labeling. Anal. Chem. 85, 8618–8625 [DOI] [PubMed] [Google Scholar]
- 77. Calabrese A. N., Ault J. R., Radford S. E., and Ashcroft A. E. (2015) Using hydroxyl radical footprinting to explore the free energy landscape of protein folding. Methods 89, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Cravello L., Lascoux D., and Forest E. (2003) Use of different proteases working in acidic conditions to improve sequence coverage and resolution in hydrogen/deuterium exchange of large proteins. Rapid Commun. Mass Spectrom. 17, 2387–2393 [DOI] [PubMed] [Google Scholar]
- 79. Rand K. D., Zehl M., Jensen O. N., and Jorgensen T. J. (2009) Protein hydrogen exchange measured at single-residue resolution by electron transfer dissociation mass spectrometry. Anal. Chem. 81, 5577–5584 [DOI] [PubMed] [Google Scholar]
- 80. Kan Z. Y., Walters B. T., Mayne L., and Englander S. W. (2013) Protein hydrogen exchange at residue resolution by proteolytic fragmentation mass spectrometry analysis. Proc. Natl. Acad. Sci. U.S.A. 110, 16438–16443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Klinger A. L., Kiselar J., Ilchenko S., Komatsu H., Chance M. R., and Axelsen P. H. (2014) A synchrotron-based hydroxyl radical footprinting analysis of amyloid fibrils and prefibrillar intermediates with residue-specific resolution. Biochemistry 53, 7724–7734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Jones L. M., J B. S., J A. C., and Gross M. L. (2011) Fast photochemical oxidation of proteins for epitope mapping. Anal. Chem. 83, 7657–7661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kaur P., Kiselar J., Yang S., and Chance M. R. (2015) Quantitative protein topography analysis and high-resolution structure prediction using hydroxyl radical labeling and tandem-ion mass spectrometry (MS). Mol. Cell. Proteomics 14, 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kaur P., Tomechko S. E., Kiselar J., Shi W., Deperalta G., Wecksler A. T., Gokulrangan G., Ling V., and Chance M. R. (2015) Characterizing monoclonal antibody structure by carboxyl group footprinting. MAbs 7, 540–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhang H., Shen W., Rempel D., Monsey J., Vidavsky I., Gross M. L., and Bose R. (2011) Carboxyl-group footprinting maps the dimerization interface and phosphorylation-induced conformational changes of a membrane-associated tyrosine kinase. Mol. Cell. Proteomics 10, M110 005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhang B., Rempel D. L., and Gross M. L. (2016) Protein Footprinting by Carbenes on a Fast Photochemical Oxidation of Proteins (FPOP) Platform. J. Am. Soc. Mass Spectrom. 27, 552–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Manzi L., Barrow A. S., Scott D., Layfield R., Wright T. G., Moses J. E., and Oldham N. J. (2016) Carbene footprinting accurately maps binding sites in protein-ligand and protein-protein interactions. Nat. Commun. 7, 13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Huang W., Ravikumar K. M., Chance M. R., and Yang S. (2015) Quantitative mapping of protein structure by hydroxyl radical footprinting-mediated structural mass spectrometry: a protection factor analysis. Biophys. J. 108, 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ephrussi A., Church G. M., Tonegawa S., and Gilbert W. (1985) B lineage–specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science 227, 134–140 [DOI] [PubMed] [Google Scholar]
- 90. Giniger E., Varnum S. M., and Ptashne M. (1985) Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell 40, 767–774 [DOI] [PubMed] [Google Scholar]
- 91. Mueller P. R., and Wold B. (1989) In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science 246, 780–786 [DOI] [PubMed] [Google Scholar]
- 92. Bertrand E., Fromont-Racine M., Pictet R., and Grange T. (1993) Visualization of the interaction of a regulatory protein with RNA in vivo. Proc. Natl. Acad. Sci. U.S.A. 90, 3496–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kim H., Abeysirigunawarden S. C., Chen K., Mayerle M., Ragunathan K., Luthey-Schulten Z., Ha T., and Woodson S. A. (2014) Protein-guided RNA dynamics during early ribosome assembly. Nature 506, 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Clatterbuck Soper S. F., Dator R. P., Limbach P. A., and Woodson S. A. (2013) In vivo X-ray footprinting of pre-30S ribosomes reveals chaperone-dependent remodeling of late assembly intermediates. Mol. Cell 52, 506–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hulscher R. M., Bohon J., Rappé M. C., Gupta S., D'Mello R., Sullivan M., Ralston C. Y., Chance M. R., and Woodson S. A. (2016) Probing the structure of ribosome assembly intermediates in vivo using DMS and hydroxyl radical footprinting. Methods 103, 49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zhang H., Tang X., Munske G. R., Tolic N., Anderson G. A., and Bruce J. E. (2009) Identification of protein-protein interactions and topologies in living cells with chemical cross-linking and mass spectrometry. Mol. Cell. Proteomics 8, 409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zheng C., Yang L., Hoopmann M. R., Eng J. K., Tang X., Weisbrod C. R., and Bruce J. E. (2011) Cross-linking measurements of in vivo protein complex topologies. Mol. Cell. Proteomics 10, M110 006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yang L., Zheng C., Weisbrod C. R., Tang X., Munske G. R., Hoopmann M. R., Eng J. K., and Bruce J. E. (2012) In vivo application of photocleavable protein interaction reporter technology. J. Proteome Res. 11, 1027–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Weisbrod C. R., Chavez J. D., Eng J. K., Yang L., Zheng C., and Bruce J. E. (2013) In vivo protein interaction network identified with a novel real-time cross-linked peptide identification strategy. J. Proteome Res. 12, 1569–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Espino J. A., Mali V. S., and Jones L. M. (2015) In Cell Footprinting Coupled with Mass Spectrometry for the Structural Analysis of Proteins in Live Cells. Anal. Chem. 87, 7971–7978 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.