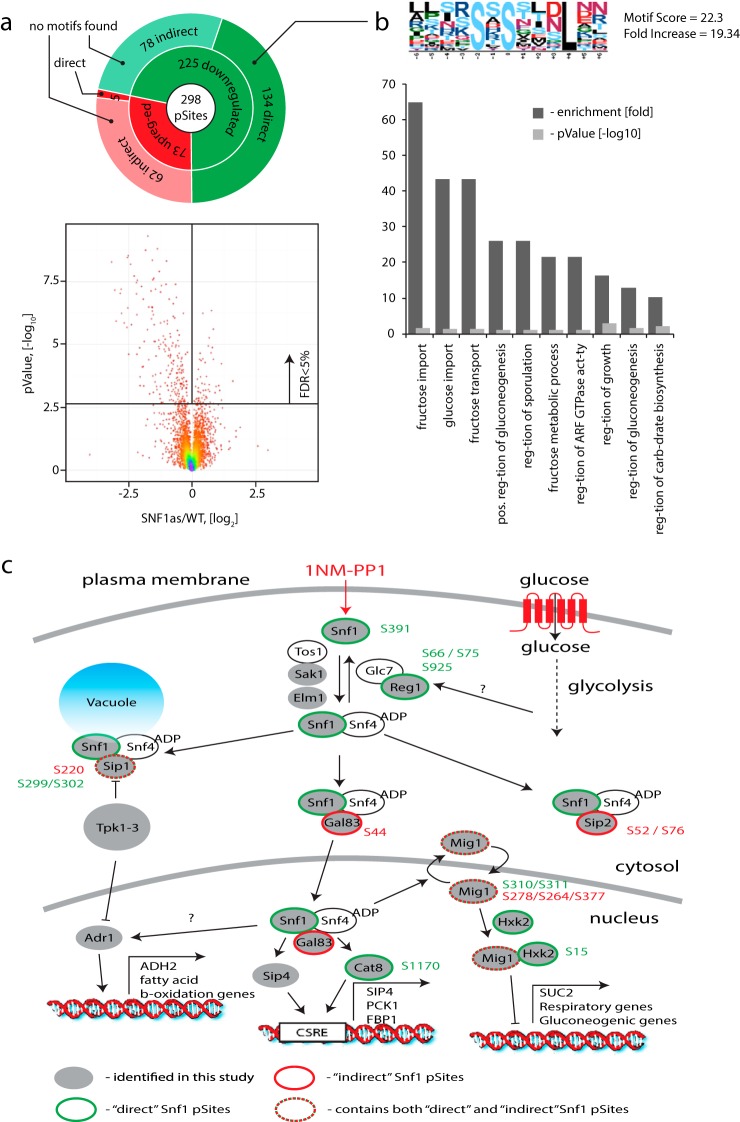
Fig. 6.
Identification of direct Snf1 substrates. (a) Application of global phosphorylation profiles and machine learning to identify direct targets of the Snf1 kinase. After removal of 1NM-PP1 inhibitor off-target phosphorylation events, significant regulation was observed for ∼ 5% of all detected sites with a strong prevalence of dephosphorylation. SVM model allowed separation of direct (134) and indirect (78) Snf1 targets among all dephosphorylated targets. (b) MotifX analysis revealed enrichment of a sequence motif where the phosphorylated residue was followed by leucine in the +4 position for direct Snf1 substrates. No significantly enriched motifs were obtained in a similar analysis of indirect Snf1 targets. Gene ontology analysis revealed enrichment of terms associated with sugar metabolism and regulation of cell growth. (c) Many known components of the local Snf1 signaling network were identified as either direct or indirect targets.