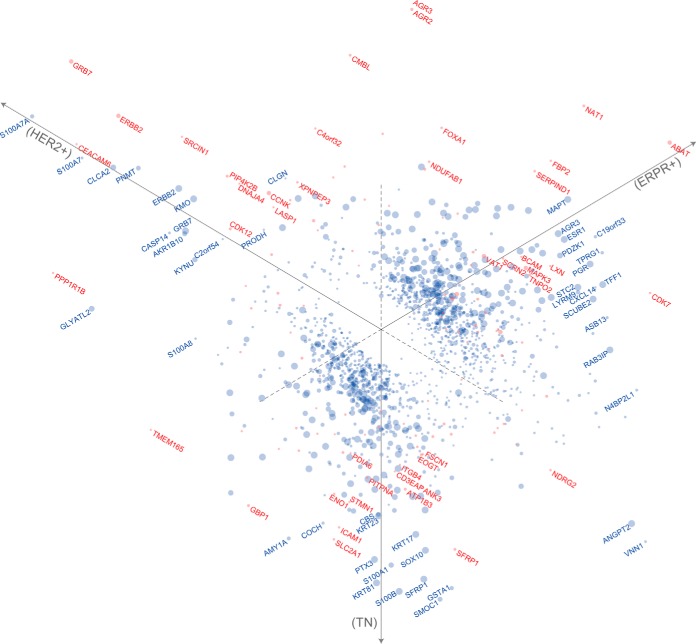
Fig. 5.
Three-way comparisons of breast cancer subtypes from CPTAC and MPIB proteomics data. Diffacto summarized differential proteins from CPTAC (blue) and MPIB (red) data sets. The differentially expressed proteins were more comprehensive and subtype-specific in the former data set, because of its larger the sample size and higher sequence coverage. Particularly, for the triple-negative subtype, which has been characterized by the high abundances of basal-like cytokeratins (56), SMOC1, S100B, GSTA1, SFRP1, S100A1, PTX3, SOX10, ANGPT2, COCH, as well as many other known markers such as SYNM1 (57), MFI2 (58), NDRG2 (59), CRYAB (60), and PLA2G4A (61) were found on top of the list of differential regulated proteins in the CPTAC data (supplemental Table S3). Markers are located on the scatter plot based on the fold changes relative to the three subtypes (axes) in log-scale. Marker radius is proportional to the q value (FDRMC) in negative logarithm scale.