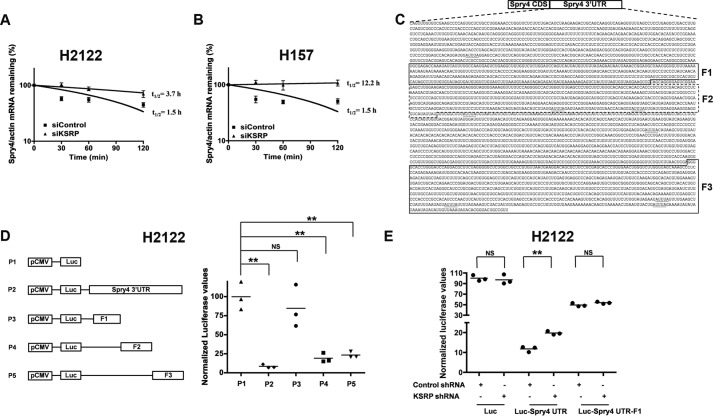
Figure 6.
KSRP promotes post-transcriptional destabilization of Spry4 transcripts. A and B, H2122 (A) and H157 (B) cells were transfected with either control or KSRP siRNAs followed by treatment with actinomycin D for the indicated periods of time. Spry4 and actin mRNA levels were quantified using qPCR, and the amounts of Spry4/actin mRNA remaining were plotted on a semilogarithmic scale. Error bars represent mean ± S.E. **, p < 0.01, ANOVA. C, schematic representation of the Spry4 mRNA with 3′-UTR displaying class I AREs. D, H2122 cells were transiently transfected with pcDNA-Luc or pcDNA-Luc with either full-length Spry4 3′-UTR (bases 1160–4961) or Spry4 3′-UTR truncations F1 (bases 2012–2650), F2 (bases 2651–3006), and F3 (bases 4072–4941). Later, the luciferase activities in the cell lysates were assayed using a luminometer, and normalized luciferase values are displayed in the graph. **, p < 0.01, ANOVA. E, H2122 cells were co-transfected with the indicated plasmids. Later, the luciferase activities in the cell lysates were assayed using a luminometer, and normalized luciferase values are displayed in the graph. **, p < 0.01, ANOVA. NS, not significant.