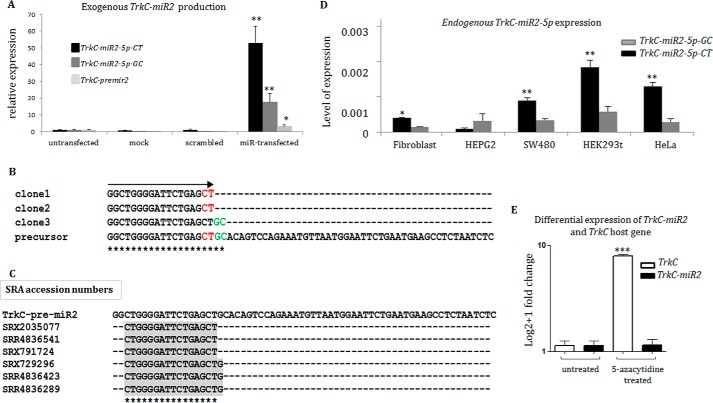
Figure 2.
TrkC-miR2–5p sequence characterization and evidence for an independent promoter. A, overexpression of TrkC-premir2 in SW480 cells and detection of predicted TrkC-miR2–5p mature forms (-CT and -GC isomiRs) by RT-qPCR. Note that TrkC-miR2–5p-CT isomiR is dominantly produced in the transfected cells. B, sequencing results of three TA vector clones containing TrkC-miR2–5p sequences created in A. Clones 1 and 2 show the sequence of TrkC-miR2–5p-CT isomiR, and clone 3 shows the sequence of TrkC-miR2–5p-GC isomiR. TrkC-miR2 sequences are aligned with the sequence of their precursor. The significance of asterisks in panels C and B is the complete matching of aligned sequences at each position. C, shown is the presence of TrkC-miR2 in the SRA data. Three reads for detected TrkC-miR2-5p-CT and TrkC-miR2-5p-CTG isomiRs are shown. However, no read for TrkC-miR2-5p-GC was detected in the SRA data set. D, shown is the expression level of TrkC-miR2–5p-CT and -GC isomiRs in several human cell lines. TrkC-miR2–5p-CT relative expression was higher than TrkC-miR2–5p-GC isomiR in most of these cell lines. E, treatment of HCT-116 cells with epidrug resulted in TrkC expression elevation (100 times). However, it did not have such an effect on TrkC-miR2–5p expression level. Error bars indicate S.D. of duplicate experiments. U48 RNA and GAPDH were used as internal controls for the amplifications. *, p < 0.05; **, p < 0.01; ***, p < 0.001.