FIGURE 3.
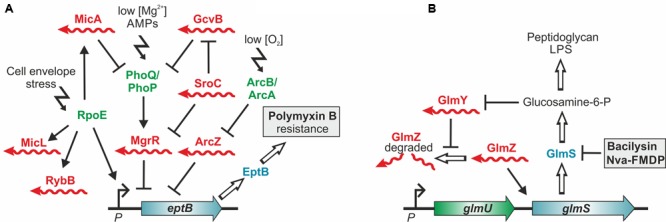
Control of antibiotic resistance by trans-encoded sRNAs. (A) Control of polymyxin resistance in E. coli. Enzyme EptB provides resistance to polymyxin B by modification of LPS with phosphoethanolamine. Translation of eptB mRNA is inhibited by sRNA MgrR, which is itself repressed by base pairing with the sponge sRNA SroC. Consequently, loss of MgrR increases and loss of SroC decreases resistance to polymyxin B. In addition, eptB is repressed by sRNA ArcZ (Moon et al., 2013), whose levels are controlled by the aerobic/anaerobic-sensing ArcA–ArcB two-component system (Mandin and Gottesman, 2010). Counterintuitively, deletion of Hfq, which is required for activity of these sRNAs increases susceptibility to polymyxin B. This might be explained by a defective cell envelope stress response executed by RpoE. RpoE not only activates transcription of eptB but also of further Hfq-dependent sRNAs, which control LPS biogenesis and modification. Complexity is further increased by the fact that mgrR transcription is activated by the two-component system PhoQ/PhoP, which is repressed by sRNAs MicA and GcvB. (B) sRNA-mediated resistance to antibiotics targeting the cell wall biosynthesis enzyme GlmS. In Enterobacteriaceae small RNAs GlmY and GlmZ feedback-regulate GlmS synthesis to achieve homeostasis of the essential metabolite GlcN6P. Inhibition of GlmS by bacilysin and other antibiotics depletes GlcN6P, which is sensed by sRNA GlmY triggering its accumulation. By a mimicry mechanism GlmY counteracts degradation of the homologous sRNA GlmZ, which in turn selectively activates translation of glmS encoded within the glmUS operon. As a result, higher GlmS levels are produced compensating for its inhibition.
