FIGURE 1.
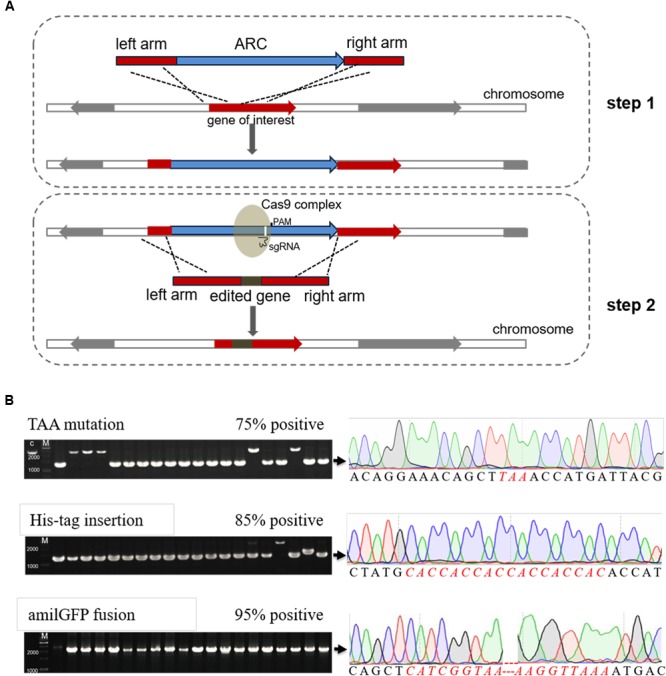
Efficient genome editing by employing both an antibiotic resistance cassette (ARC) and the CRISPR/Cas9 system. (A) Schematic chart illustrating the two-step strategy for editing non-essential genes (or any DNA sequences) in E. coli. Firstly, an ARC was inserted into the genome near to the target site through homology-directed repair (HDR). Secondly, the Cas9 complex targeting the ARC sequence cleaved the ARC, and meanwhile the ARC was replaced with the edited sequence through the second step of HDR. The target sequences could be mutated, deleted or inserted with foreign DNA sequences, and the λ-Red recombination system remarkably increased the HDR efficiency in both steps. (B) Verification of the edited mutants by both colony PCR (left) and Sanger DNA sequencing (right). Correct sizes are indicated at the end of arrows, and the positive rates are labeled. The PCR products that formed positive clones were then randomly selected for Sanger DNA sequencing analysis. Detailed sequences can be found in Supplementary Figure S3, and the inserted amilGFP sequence contained its own promoter. M, GeneRuler 1-kb DNA ladder (ThermoFisher Scientific).
