Fig. 1.
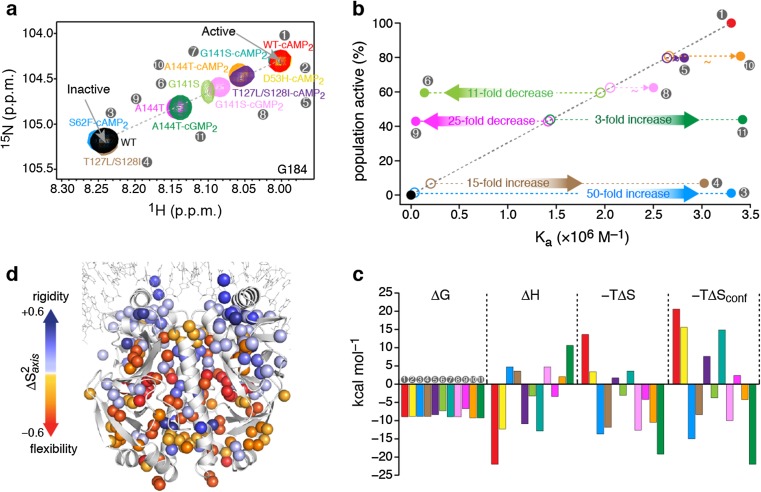
a Overlaid 1H–15N heteronuclear single quantum coherence (HSQC) spectra of the CAP variants, showing the resonance of the G184 amino acid, a residue located in the DNA recognition helix. The chemical shift of apo WT-CAP (labeled WT) indicates the inactive conformation of the DBD, whereas the chemical shift of WT-CAP-cAMP2 indicates the active conformation of the DBD. Each CAP variant is denoted by a number (gray circles) and color, which are the same in each figure panel. b The relative population of each CAP variant in the active DBD state as a function of the variant’s theoretical (open circles) and experimentally determined (filled circles) affinity for DNA. c Thermodynamic components of the binding of the CAP variants to DNA. d Effect of DNA binding on the methyl order parameters, S 2 axis, of CAP-T127L/S128I. Figure reproduced from Tzeng and Kalodimos 2012