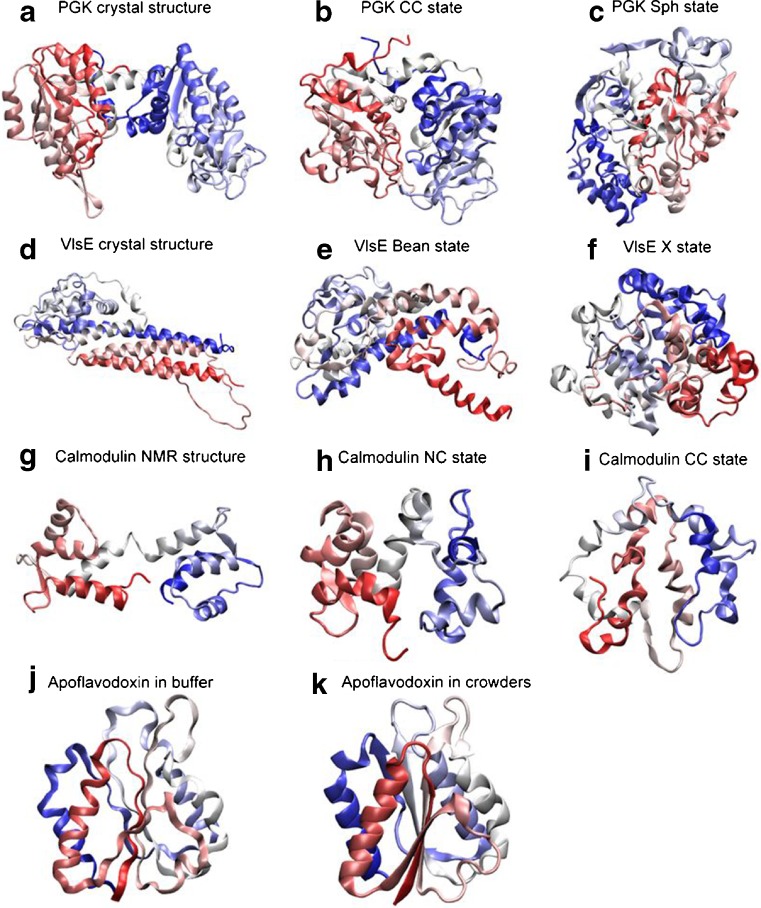
Fig. 2.
Cartoon representations of folded-state changes (with respect to crystal or NMR structures) found in silico for four proteins (PGK, Dhar et al. 2010; VlsE, Homouz et al. 2008; calmodulin, CaM, Homouz et al. 2009a; apoflavodoxin, Stagg et al. 2007) in the presence of macromolecular crowding. For each row, from left to right, the crowding level is increased. The shapes of the proteins are characterized by the shape parameter S (S ranges from −0.25 to 2: S < 0 is oblate, S = 0 is spherical, and S > 0 is prolate) and the asphericity Δ (Δ ranges from 0 to 1: Δ = 0 is sphere and Δ = 1 is rod). For CaM, due to the fact that the N- and C- lobes have different stability, besides Δ, an overlap function χ is used to characterize structures. χ = 0 indicates that a structure is identical to its NMR structure. χN is the overlap function for the N-lobe of CaM. Cartoon representations are colored from the N-terminus (red) to the C-terminus (blue). a crystal structure of PGK (which is extended; S = 0.26, Δ = 0.26); b collapsed crystal (CC) structure of PGK in crowders (S = 0.05, Δ = 0.1); c spherical compact (Sph) structure of PGK in crowders (S = 0.01, Δ = 0.02); d crystal structure of VlsE (which has an american football shape; S = 0.34, Δ = 0.31); e bean-like structure (Bean) of VlsE in crowders (S = 0.14, Δ = 0.17); f spherical structure (X) of VlsE in crowders (S = 0.02, Δ = 0.02); g NMR structure of CaM (which has a dumbbell-like shape; 0.2 < Δ < 0.4); h ‘native collapsed’ (NC) structure of CaM in crowders (this ensemble has a structurally native-like N-lobe and an unfolded C-lobe that is collapsed over the N-lobe; 0.0 < χN < 0.1 and 0.0 < Δ < 0.2); i completely collapsed (CC) structure of CaM in crowders (here, both N- and C- lobes have collapsed; 0.2 < χN < 0.4 and 0.0 < Δ < 0.2); j structure of apoflavodoxin in buffer (radius of gyration Rg of 15.40 Å) and, k structure of apoflavodoxin in crowders (Rg reduces to 15.35 Å)