Abstract
The division of Escherichia coli is an essential process strictly regulated in time and space. It requires the association of FtsZ with other proteins to assemble a dynamic ring during septation, forming part of the functionally active division machinery, the divisome. FtsZ reversibly interacts with FtsA and ZipA at the cytoplasmic membrane to form a proto-ring, the first molecular assembly of the divisome, which is ultimately joined by the rest of the division-specific proteins. In this review we summarize the quantitative approaches used to study the activity, interactions, and assembly properties of FtsZ under well-defined solution conditions, with the aim of furthering our understanding of how the behavior of FtsZ is controlled by nucleotides and physiological ligands. The modulation of the association and assembly properties of FtsZ by excluded-volume effects, reproducing in part the natural crowded environment in which this protein has evolved to function, will be described. The subsequent studies on the reactivity of FtsZ in membrane-like systems using biochemical, biophysical, and imaging technologies are reported. Finally, we discuss the experimental challenges to be met to achieve construction of the minimum protein set needed to initiate bacterial division, without cells, in a cell-like compartment. This integrated approach, combining quantitative and synthetic strategies, will help to support (or dismiss) conclusions already derived from cellular and molecular analysis and to complete our understanding on how bacterial division works.
Keywords: Physical biochemistry, Mechanistic biochemistry, Synthetic biology, Protein–protein interactions, Protein–membrane interactions, Biophysical methods
Introduction
This article is not a comprehensive description of the advances made in our understanding of FtsZ and its central role in bacterial division—a subject covered in recent reviews (Adams and Errington 2009; Egan and Vollmer 2013; Erickson et al. 2010; Lutkenhaus et al. 2012; Mingorance et al. 2010). Rather, it reflects our own views and reflections on the field. We have focused on the quantitative analysis of the activities and association/assembly properties of FtsZ, their modulation by ligands and excluded-volume effects due to crowding, and their study in different membrane-like systems. This knowledge will complete our understanding of the functional energetics of macromolecular interactions involved in bacterial division and will contribute to defining the precise conditions to build, with a minimum set of elements, molecular assemblies that reproduce (at least in part) an essential biological process in the test tube. This integrative approach, which given the current trends seems a feasible task, reflects the teachings of Allen Minton during our long-term (personal and scientific) relationship.
The bacterial (Escherichia coli) division machinery
The components of the multi-protein machinery that mediates bacterial division reversibly interact during the assembly of a division ring at midcell to drive cytokinesis (Adams and Errington 2009; Erickson et al. 2010; Margolin 2000; Mingorance et al. 2010). The ring is a highly dynamic entity composed of at least ten proteins, most of which are integral membrane proteins, whose structural organization is not yet precisely established. They form part of the complete macromolecular assembly capable of effecting division in the cell, namely, the divisome (Nanninga 1998; Vicente 2013), which also contains periplasmic, outer membrane and cell envelope-synthesizing elements (Egan and Vollmer 2013; Vicente and Rico 2006).
In E. coli, division is initiated by the concerted association of three proteins, i.e., FtsZ, FtsA, and ZipA, at the cytoplasmic membrane to form the first macromolecular complex of the divisome, the proto-ring, which in turn acts as a scaffold where the remaining division proteins are incorporated (Geissler et al. 2007; Pichoff et al. 2012; Vicente and Rico 2006) (Fig. 1). Among the components of the proto-ring, the best characterized is FtsZ, a GTPase widely conserved across prokaryotic organisms and structurally related to tubulin, a major component of the eukaryotic cytoskeleton. FtsZ, the most abundant element of the division ring, is an assembling machine capable of forming single-stranded filaments that further associate into higher order structures. These assembly reactions are considered to be crucial to the division function in which force-generating mechanisms are invoked (Erickson et al. 2010; Mingorance et al. 2010). The precise structural organization of the Z-ring has not yet been identified, and the correlation of the microscopic images of the division rings with a particular FtsZ polymeric entity has not yet been established. These difficulties may relate in part with the plasticity of these assemblies and with the dynamic nature of the reversible interactions between division elements (Strauss et al. 2012; Vicente 2013). The application of high-resolution imaging techniques is expected to produce a better reconstruction of the Z-ring, revealing details on its molecular architecture in the cell.
Fig. 1.
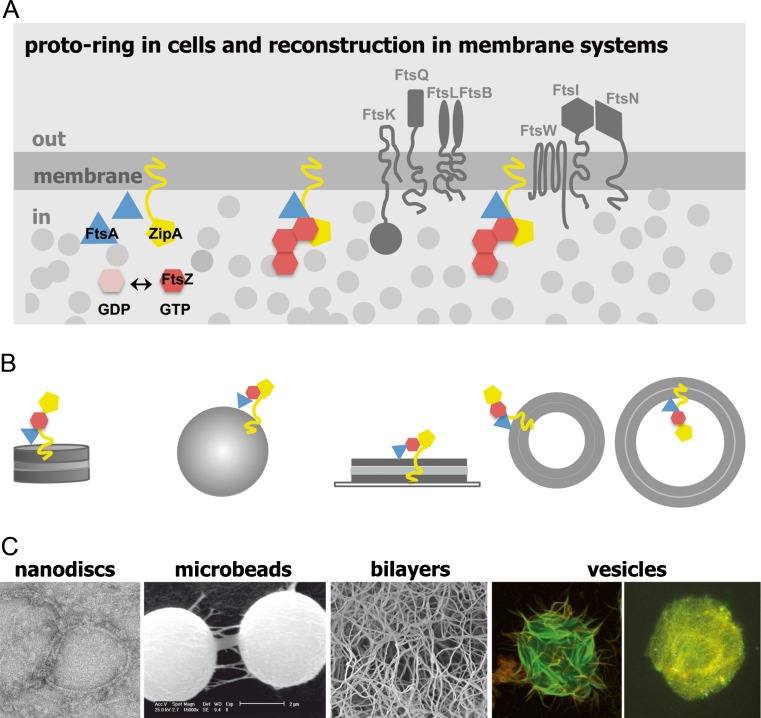
Proto-ring in cells and in reconstructed membrane systems. a. Diagram illustrating the position of essential divisome elements in Escherichia coli. Proto-ring elements are colored, while other divisome proteins are shown in dark gray. Light-gray circles represent the macromolecules contributing to the crowding of the cytoplasm. b. Reconstruction of proto-ring in membrane systems. Vesicles show the two alternative orientations of the reconstructed proto-ring relative to the vesicle lumen. c. Visualization of guanosine triphosphate (GTP)-induced FtsZ polymers reconstituted in different membrane systems. From left to right Transmission electron microscopy micrograph of ZipA-containing nanodiscs, scanning electron microscopy (SEM) micrograph of microbeads coated with native inner membrane, SEM micrograph showing FtsZ polymers growing in a three-dimensional network on a ZipA-containing lipid bilayer, confocal microscopy images of giant unilamellar vesicles with FtsZ assembled outside or inside the vesicles. Adapted from Hernández-Rocamora et al. (2012a), Jiménez et al. (2011), and Martos et al. (2012a), with permission
FtsZ is anchored to the cytoplasmic membrane through the interaction of its carboxy-terminal region with the other proto-ring proteins, ZipA or FtsA (Haney et al. 2001). Apparently either of the latter can support the attachment of FtsZ, but no localization of FtsZ at the membrane occurs if both are absent (Geissler et al. 2007; Pichoff et al. 2012). The amphitropic protein FtsA is considered to be a member of the actin family and contains a short amphipathic helix that is thought to mediate its association with the membrane (Pichoff and Lutkenhaus 2005). It has recently been shown that FtsA associates into filaments (Martos et al. 2012b), in this case, in the presence of lipid structures; FtsA assembly has also been observed in other bacterial cells (Krupka et al. 2012; Szwedziak et al. 2012). ZipA contains a short amino-terminal region integrated in the membrane and connected to the carboxy-terminal FtsZ-interacting domain by a flexible, unstructured linker region (Ohashi et al. 2002).
The positioning of the E. coli division ring is regulated by two control mechanisms, namely, the Min system and nucleoid occlusion, which work together to inhibit ring formation in the wrong places. The Min system comprises three proteins (Min C, D, and E); the pole-to-pole oscillatory behavior of the interacting MinCD (putative inhibitors of the FtsZ ring) directed by a MinE ring creates a gradient of the Min inhibitors such that their local concentration is lowest at midcell, favoring FtsZ assembly in this region (Loose et al. 2008). Nucleoid occlusion is an additional mechanism of regulation to avoid septation at places occupied by the non-divided bacterial nucleoid (Wu and Errington 2012), a soft colloidal entity that exerts its inhibitory control through both nonspecific excluded-volume and electrostatic effects and the specific action of SlmA, a DNA-binding protein recognizing specific sequences throughout the nucleoid that inhibits FtsZ ring assembly in the vicinity of bulk chromosomal DNA (Tonthat et al. 2011).
The bacterial cytoplasm is a crowded environment
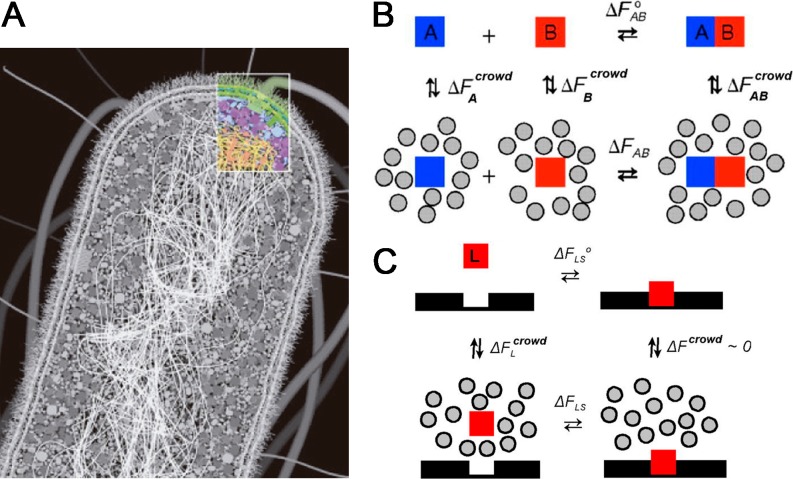
The assembly of functional division complexes in vitro requires environments in which the natural crowding conditions are carefully reproduced. There are major differences between the intact bacterial cytoplasm and conventional solutions studied in the laboratory, including large differences in total protein concentration and in the fraction of total volume that lies adjacent to the surface of either a membrane or a fibrous structural element. It is estimated that around 25–30 % of the bacterial cytoplasm volume is occupied by macromolecules (among them approx. 200–300 mg/ml protein, approx. 100 mg/ml RNA, and approx. 15 mg/ml DNA; Vendeville et al. 2011), while no single macromolecule needs to be highly concentrated (Fig. 2); for example, the estimated average concentration of FtsZ, one of the most abundant cell division proteins, is 0.1–0.4 mg/ml (Lu et al. 1998; Rueda et al. 2003).
Fig. 2.
The crowded cell interior: biological organization and energetic consequences. a. Diagram illustrating the overall architecture of the E. coli cell. The double-layered cell wall and the two membranes comprise the structural zone of the cell (Spitzer and Poolman 2009). The crowded interior is roughly separated in two zones—a soluble region containing most of the proteins and ribosomes (the metabolic zone) and the nucleoid region filled mostly by the DNA genome (the replication zone). Reprinted with permission from Goodsell (2010). b, c Thermodynamic cycles illustrating linkage between free energy of transfer of reactants and products from a dilute solution to a crowded medium and standard free energy of association in solution (b) and site-binding (c). Figure modified from Zhou et al. (2008)
Although much of the chemistry associated with bacterial division reactions takes place in environments containing a substantial volume fraction of macromolecules, local composition may vary widely within the bacterial cell, and macromolecules inside the cell seem to be transiently clustered in functional and structural networks rather than homogeneously distributed. Preferential location (or exclusion) of a certain component arises from increased heterogeneity and the probable presence of additional weak nonspecific attractive and repulsive associations with background molecules or large structures. For example, FtsZ, in addition to being subjected to pure volume-exclusion effects, will also be subjected to the influence of the local microenvironments where it can be found during its life cycle: (1) it is located in the immediate vicinity of the nucleoid, a highly charged soft supramolecular entity that occupies approximately 15 % of the bacterial interior, where FtsZ will encounter a high local concentration of nucleic acid and charges; (2) it is also found in the vicinity of the inner plasma membrane, to which FtsZ is anchored as part of the division ring, which provides high local concentration of membrane proteins and phospholipids. It is expected that the relative contributions of crowding, confinement, and adsorption to FtsZ reactivity will be different within each of these microenvironments; however, despite such complexity, it is true that background interactions arising from steric repulsion in volume-occupied cell-like environments will have to be taken into account, independently of the presence or absence of other types of interactions (Minton 2006; Phillip and Schreiber 2013; Zhou 2013).
Because of its generality, special attention has been paid to the consequences and implications of excluded volume effects due to crowding, as they arise from the mutual impenetrability of macromolecules. A fundamental chemical consequence of volume exclusion is that provides a generalized nonspecific force to facilitate processes leading to a reduction in excluded-volume, namely, macromolecular compaction and association (Ellis 2001; Minton 2000, 2001) (Fig. 2). It has been shown that larger and less compact macromolecules are more sensitive to excluded-volume effects than smaller and more compact species, leading, under certain circumstances, to the spatial self-organization of fibers (Herzfeld 2004). Many of these effects have been observed experimentally [reviewed in Minton (2006) and Zhou et al. (2008)]. Simple statistical–thermodynamic theories, based on coarse-grained models, usually provide reliable predictions of qualitative effects and, in favorable cases, can provide reasonable semi-quantitative calculations of the magnitude of such effects (Hall and Minton 2003; Zhou et al. 2008).
The potential implications of additional specific and nonspecific interactions, other than volume exclusion, on the energetics and dynamics of macromolecular reactions in crowded environments have been raised since the first days of crowding studies (Minton 1983, 1990). Both experimental and in silico studies indicate that, under certain conditions and environments, crowding can modulate the stability and associations of some proteins through effects that are smaller than what would be predicted by the excluded-volume theory, or even the opposite. To explain this discrepancy, additional attractive interactions between background molecules and the reactants studied have been invoked; these compensate (to a greater or lesser degree) for the repulsive steric interaction due to volume exclusion (Elcock 2010; Jiao et al. 2010; Minton 2013; Minton and Wilf 1981; Phillip and Schreiber 2013). While excluded-volume effects are ubiquitous, the impact of compensating attractive interactions is highly variable and system-dependent as they vary with the chemical nature of the interacting species and the type of reactions studied. We have to date focused upon excluded-volume effects since the behavior of the proteins that we have studied in crowded model solutions may be satisfactorily described qualitatively and often quantitatively by excluded-volume models (Alcorlo et al. 2009; González et al. 2003; Jimenez et al. 2007; Rivas et al. 1999, 2001; Zorrilla et al. 2004a). Because the probability that interactions other than excluded-volume contribute significantly to total crowding effects increases as the complexity and heterogeneity of the crowded medium increases, our future research plans call for including other parameters modulating the crowding effects, such as temperature, as temperature dependence has been shown to be diagnostic for the presence and magnitude of attractive interactions (Jiao et al. 2010; Minton 2012, 2013). We believe that experimental and theoretical studies of systems of controlled but increasingly complex and progressively more cell-like composition are a necessary prerequisite to understanding macromolecular behavior in a cell.
FtsZ activities, associations, and assembly: a quantitative biochemistry approach
A prerequisite to reconstruct minimal proto-ring assemblies in the test tube is to quantitatively understand and control the association and assembly behavior of FtsZ, processes which are allegedly relevant for the central role that FtsZ plays in division. Although as with tubulin these reactions depend on magnesium and guanine nucleotides and involve guanosine triphosphate (GTP) hydrolysis (Erickson et al. 2010; Mingorance et al. 2010), the control of E. coli FtsZ assembly by ligands presents certain peculiarities. While the binding of nucleotides to FtsZ does not require magnesium, hydrolysis and dissociation of bound GTP does depend upon this divalent cation, behavior more related with that observed for GTPases of the Rho family (Erickson et al. 2010; Mingorance et al. 2010; Zhang et al. 2000; Rivas and co-workers, unpublished results). The protomeric unit of FtsZ is a monomer, which binds magnesium with low affinity (Rivas and co-workers, unpublished results) while tubulin contains a high-affinity magnesium-binding site that controls the structural stability of tubulin heterodimer (Menendez et al. 1998).
The GTP-induced FtsZ polymerization to form single-stranded protofilaments in an apparently cooperative manner can ultimately lead to a variety of supramolecular structures. Flexible multi-stranded fibers, circles, ribbons, bundles and toroids (Adams and Errington 2009; Erickson et al. 2010; González et al. 2003; Mingorance et al. 2010; Popp et al. 2009) are observed depending on solution conditions (buffer composition, protein concentration, surface effects, nonspecific effects of macromolecular additives, specific effects of other cell division proteins). This polymorphism is a reflection of the structural plasticity of FtsZ polymers, which are highly dynamic both in vivo (Stricker et al. 2002) and in vitro (Chen and Erickson 2005). Large changes in the geometry of the fibers would require very small free energy perturbation of the intermolecular interface (González et al. 2005). Experiments on FtsZ polymerization are particularly challenging because of the dynamic character of polymers, which is linked to the GTPase activity of the protein, and special procedures are required to maintain polymer stability throughout the duration of the experiments (González et al. 2003). All these may explain, in part, the controversy which has emerged in recent years regarding the GTP-dependent mechanism of FtsZ polymerization (Erickson et al. 2010; Mingorance et al. 2010). Current biochemical and biophysical research on FtsZ is aimed at describing the polymerization and force-generating mechanisms and evaluating the roles of nucleotide exchange and hydrolysis; the results of such studies will help to determine the precise role of FtsZ in division (Mingorance et al. 2010; Salvarelli et al. 2011).
A good description of these ligand-linked FtsZ interactions requires knowing how changes in the concentrations of proteins and physiological ligands influence the association schemes and the relative abundance of the FtsZ species involved. We have obtained this information by using complementary biophysical approaches that combine analytical ultracentrifugation, static/dynamic light scattering, and fluorescence spectroscopy methods; this strategy has provided a thorough and robust description of these associations and assembly reactions. The choice of the experimental approach used has been based upon the processes being studied, either associations of the guanosine diphosphate (GDP)–FtsZ forms or the assembly of FtsZ in the presence of GTP or analogs.
Mg2+-linked non-cooperative self-association of FtsZ monomers in the presence of GDP
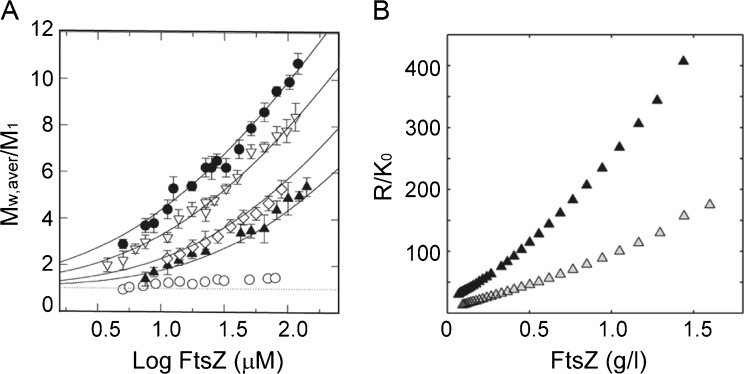
FtsZ monomers, equilibrated in neutral pH buffers containing GDP, were found to self-associate through a Mg2+-linked non-cooperative process as revealed by the composition-dependence of the molar mass of FtsZ, measured by sedimentation equilibrium over a broad range of protein and ligand concentrations (Martos et al. 2010; Rivas et al. 2000) (Fig. 3). The mechanism of oligomer formation is best described as an indefinite self-association reaction coupled to the binding of a molecule of Mg2+ for each FtsZ added to the growing oligomer. The tendency of FtsZ to self-associate decreases as the buffer ionic strength increases, with 14-mers and hexamers being the larger class of oligomers detectable at 0.05 and 0.5 M K+, respectively. The self-association behavior, and its dependence on solution conditions, was characterized by composition-gradient static light scattering (CG-SLS), a technique that greatly facilitates the rapid (order of minutes) acquisition of continuous light scattering data from a solution whose composition is being varied with time in a controlled manner (Attri and Minton 2005a, b). The resulting time- and composition-dependent scattering profiles were quantitatively best described by the same molecular schemes (adapted to model the composition dependence) that accounted for the sedimentation equilibrium data (Fig. 3). The sedimentation coefficient distributions calculated from sedimentation velocity experiments were compatible with the self-association features of FtsZ in its GDP form mentioned above (Martos et al. 2010; Rivas et al. 2000) and their modulation by magnesium and potassium ions. These findings suggest that, at physiologically relevant concentrations of these ligands in E. coli (free Mg2+ approx. 1–2 mM, intracellular K+ approx. 0.1–1 M; Chang and Cronan 1986; Record et al. 1998), GDP–FtsZ has a weak tendency to form oligomers in solution.
Fig. 3.
Self-association of FtsZ in guanosine diphosphate (GDP)-containing solutions. a Mg2+ dependence of the state of association of FtsZ (M w,aver/M 1) upon protein concentration. Magnesium concentrations are approx. 1 μM (open circles), 1 mM (solid triangles), 2 mM (open diamonds), 5 mM (open triangles) and 10 mM (solid circles). Solid lines are the corresponding best fits of indefinite self-association functions (Rivas et al. 2000). b Concentration dependence of 90° scattering of FtsZ with 5 mM magnesium at 500 (gray triangles) and 100 mM KCl (black triangles). Adapted from Rivas et al. (2000)
Mg2+-linked concerted polymerization of FtsZ in the presence of GTP or analogs
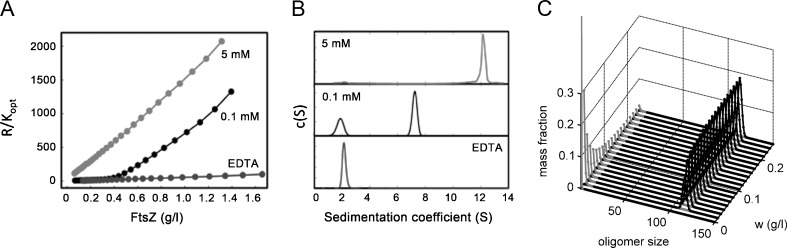
The sedimentation coefficient distributions of FtsZ in the presence of constantly replenished GTP (González et al. 2003) were measured as a function of protein and Mg2+ concentrations in close to physiological buffer solutions (neutral pH, 0.5 M K+) under steady-state conditions (González et al. 2005; Monterroso et al. 2012a). As the concentration of protein increases, FtsZ undergoes a concerted transition between a paucidisperse distribution of slowly sedimenting species (2–4 S) and a narrow size distribution of rapid species (12–13 S). The peak size of this distribution varies with the concentration of monovalent and divalent ions, but the concerted nature of the transition does not (Fig. 4). Any further increase in protein concentration increases the population of high molecular weight species, but not the mean size or dispersity, suggesting a second-order phase transition (González et al. 2005; Monterroso et al. 2012a). The concentration dependence of static light scattering (measured by CG-SLS) and diffusion coefficients (measured by fluorescence correlation spectroscopy and dynamic light scattering) independently verified the cooperative appearance of a narrow size distribution of oligomeric species providing, in addition, the size of these species, 100 ± 20 FtsZ monomers, at a millimolar Mg2+ concentration (Monterroso et al. 2012a). Parallel experiments on solutions where polymerization was triggered by GMPCPP, a slowly hydrolyzable analog of GTP, likewise indicated the concerted formation of a narrow size distribution of stable oligomers with a larger average mass, 150 ± 20 FtsZ monomers. The closely similar behavior of FtsZ in the presence of GTP or GMPCPP might be evidence of a common association scheme.
Fig. 4.
Polymerization of FtsZ in GTP-containing solutions. a. Concentration dependence of GTP–FtsZ static light scattering at 90°, supplemented with an enzymatic GTP regenerating system (RS) as described in (González et al. 2003), at the magnesium concentration specified in the figure or in the presence of 2 mM EDTA. Lines are meant only to guide the eye. b. Magnesium dependence of FtsZ sedimentation velocity [c(S)] in the presence of 1 mM GTP and RS. Sedimentation profiles of 1 g/l FtsZ equilibrated at the magnesium concentrations specified in the figure. c. Fractional abundance of FtsZ oligomeric species in the presence of GTP/RS and 5 mM magnesium, plotted as a function of oligomer size and total protein concentration, calculated using a nucleated isodesmic−cyclization model (Monterroso et al. 2012b). Gray Linear oligomers, black cyclic filaments. Adapted from Monterroso et al. (2012a, b)
These results have been semi-quantitatively described by a quasi-equilibrium model assuming that FtsZ self-assembles into linear fibers which, above a certain size and because of their natural curvature and flexibility, tend to form cyclic fibrils (Monterroso et al. 2012b) (Fig. 4). These species are not necessarily circles, as they are circular only in a topological sense and their actual conformation in solution may be far from planar and not even close to circular in shape; cyclization may reflect the formation of several simultaneous side-to-side contacts between FtsZ molecules at fiber termini leading to loop closure, rather than a single end-to-end contact. Electron microscopy (EM) and Cryo-EM images of curved and cyclic structures have been reported elsewhere, and recent atomic force microscopy (AFM) images obtained from solutions prepared under conditions identical to those studied in González et al. (2005) and Monterroso et al. (2012a, b) show abundant and relatively homogeneous populations of cyclic FtsZ fibrils of sizes comparable to those described above (Mateos-Gil et al. 2012).
It is cautious not to attribute physiological significance to the narrow size distribution of fibril species tentatively identified as cyclic oligomers, as there are many differences between the selected in vitro conditions and those found in the cell. The most obvious of these are the absence of other division proteins and of elements of the intracellular environment interacting with FtsZ (at least) in a nonspecific manner, probably contributing to the fine-tuning of Z-ring formation. Although these factors therefore have to be ultimately integrated in the synthetic approaches to reconstruct FtsZ-based minimal division machineries, the quantitative studies described here provide relevant information on the solution behavior of FtsZ that needs to be considered when elaborating more complex schemes describing the mechanism of FtsZ polymerization.
Dynamic self- and hetero-associations of GDP–FtsZ and ZipA in solution
FtsZ in its GDP form was found to bind with moderate affinity to sZipA (a soluble form of ZipA lacking the trans-membrane region), as measured by CG-SLS (Martos et al. 2010). Under close to physiological conditions, FtsZ monomers self-associate in a non-cooperative fashion to form short oligomers, with hexamers being the larger species detectable, while sZipA is a monomer. CG-SLS data from mixtures of FtsZ and sZipA were quantitatively best described by an association model where a molecule of sZipA binds any FtsZ oligomer with the same affinity (K a approx. 5 × 105 M−1). The binding of sZipA to FtsZ did not alter the affinity between FtsZ monomers, as the best-fit value of the affinity constant (K a approx. 6.5 × 104 M−1) was the same as that obtained in the absence of sZipA. The interaction between GDP–FtsZ and sZipA and the association scheme were confirmed by sedimentation equilibrium. These results would support the role of ZipA as a divisome element providing a dynamic membrane tethering for FtsZ.
Effect of crowding on FtsZ associations and assembly
It is reasonable to believe that a robust strategy to reconstruct functional proto-ring assemblies in minimal cell-like systems would require a quantitative understanding of the time-dependent and steady-state behavior of FtsZ oligomers and polymers under conditions that faithfully reproduce the intracellular environment. However, the state(s) of association and assembly of FtsZ in such cytomimetic media cannot be reliably inferred from the knowledge of these FtsZ properties measured in the absence of background components naturally present in the cell, which may interact in a nonspecific manner with FtsZ. Of those background interactions, we have already discussed the possibility that excluded-volume effects, due to crowding, may alter, both qualitatively and quantitatively, the reactivity of individual macromolecules (i.e. FtsZ).
Studying FtsZ association/assembly reactions in crowded media
Excluded-volume conditions are usually reproduced in the test tube by adding high concentrations of inert macromolecules, of natural or synthetic origin (referred to as crowding agents or crowders), to the system (Ellis 2001). The quantitative analysis of the effects of crowding on FtsZ-related reactions involves experimental challenges additional to those existing when such reactions are studied in dilute solution. First of all, the behavior of FtsZ species which are diluted has to be monitored and measured in the presence of much higher concentrations of crowding species. Secondly, any observed effect of crowder species on a given FtsZ reaction may be due to other causes, instead of volume exclusion, such as weak interactions between crowder and FtsZ species. Finally, the effects of non-ideality have to be taken into account to interpret the measurements.
Several biophysical methods have been used to measure crowding effects on FtsZ association and assembly reactions. Non-ideal tracer sedimentation equilibrium (NITSE) is a variation of SE that is particularly well adapted to detect and measure intermolecular interactions of dilute proteins in solutions containing high concentrations of background macromolecules (Rivas et al. 1999, 2001; Rivas and Minton 2004). This technique requires that the equilibrium concentration gradient of the protein (the tracer) can be measured independently of the gradients of the background species present in the solution. A given protein can be considered to be a tracer if it has an intrinsic or extrinsic (via labeling) signal that can be uniquely detected and if it is not perturbed by the presence of the crowder species. Theoretical and methodological advances in the analysis of SE in solutions of arbitrary concentration, taking into account thermodynamic non-ideality, allow a reliable interpretation of the composition dependence of signal-average buoyant molar mass in terms of repulsive and attractive interactions between species present at significant concentration. NITSE has provided quantitative information about nonspecific repulsive interactions between tracer protein and crowder components (Fodeke and Minton 2010; Rivas et al. 1999), self-association of tracer proteins (Rivas et al. 1999, 2001; Zorrilla et al. 2004a) and hetero-association of tracer components (Alcorlo et al. 2009) in crowded solutions.
Fluorescence spectroscopy methods are also powerful tools for detecting and measuring properties of tracer proteins in crowded solutions due to the possibility of selectively detecting labeled biomolecules. However, there are still only a few examples of the application of fluorescence-based assays to study crowding effects. Fluorescence anisotropy, which measures the decrease in the rotational motion of a fluorescent tracer protein when it associates with itself or with another macromolecule, has been used to detect the self-association of dilute proteins (Wilf and Minton 1981; Zorrilla et al. 2004b) and the formation of protein–DNA complexes in crowded solutions, and also to measure the influence of crowding agents on the critical concentration of GTP-linked FtsZ assembly (Reija et al. 2011). Fluorescence resonance energy transfer, which measures the distance between donors and acceptors attached to either two regions of the same protein or two different interacting macromolecules, has been used to detect and measure crowding-induced changes in the conformation of adenylate kinase protein (Nagarajan et al. 2011). Fluorescence correlation spectroscopy, which provides information on dynamic properties (translational diffusion) of tracer proteins from the fluorescence intensity fluctuations measured within a confocal volume, has been applied to characterize the motion of biomolecules in crowding conditions (Dix et al. 2006; Lavalette et al. 2006).
Crowding enhances the self-association of GDP–FtsZ
The addition of unrelated background proteins (bovine serum albumin and hemoglobin) at concentrations as high as 150 g/l was found to enhance the tendency of the GDP-containing form of FtsZ to self-associate, forming oligomers, as measured by NITSE (Rivas et al. 2001). The composition-dependence of the buoyant molar mass of FtsZ was quantitatively described by a model in which it is assumed that FtsZ interacts with each protein crowder via steric repulsion, with the crowder species and FtsZ monomers represented by equivalent rigid spheres and each FtsZ oligomer represented as an effective sphero-cylinder. This description of the data is in excellent agreement with the predictions of the excluded-volume theory, providing evidence that crowding can modulate, in a nonspecific manner, FtsZ self-association.
Crowding promotes the self-organization of GTP–FtsZ filaments to form dynamic polymer networks
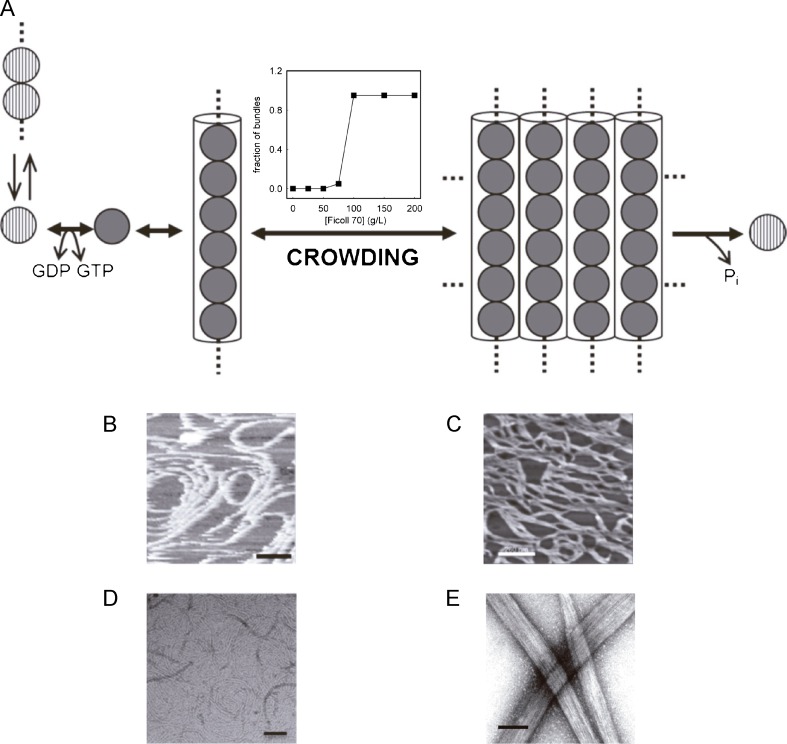
The enhancement of the rate and extent of formation of fibrous protein assemblies has been found (as expected) to be one of the major effects of volume exclusion in crowded environments on macromolecular reactions. The addition of unrelated background macromolecules (Ficoll and dextran, both with average molar mass around 70,000 Da) at concentrations as high as 150 g/l was found to promote the GTP-dependent assembly of FtsZ into dynamic multi-stranded fiber networks, as measured by turbidity and sedimentation assays and observed by EM and AFM (Fig. 5). The self-organization of FtsZ polymers induced by crowding, which is GTP-dependent, was found to retard the GTPase activity of the protein and the dynamics of FtsZ polymers when compared with the measurements obtained using FtsZ single-stranded filaments in the absence of crowding agents. In a separate study, toroidal FtsZ structures were observed in electron micrographs of FtsZ polymerized in the presence of methylcellulose or polyvinyl alcohol (Popp et al. 2009). It is not clear whether the observed effect is due to excluded-volume, as the weight concentrations of these additives that are sufficient to induce FtsZ condensation are rather low (<10 g/l) when compared with the concentration of Ficoll or dextran needed to induce a substantial condensation. It is possible that the mechanism of action of these substances to produce such interesting structures is related to solvent perturbations linked to electrostatic interactions rather than—or perhaps in addition to— excluded-volume effects due to steric repulsion. Taken together, these results lead to the proposal that in a cell-like environment under assembly-promoting conditions, the FtsZ thin filaments tend to associate, forming dynamic higher-order structures which may fit into the Z-ring. These spontaneous FtsZ assembly processes must be highly regulated in vivo by specific mechanisms to prevent the formation of the Z-ring in non-dividing cells.
Fig. 5.
Crowding effects on FtsZ self-association and assembly. a. Model for FtsZ assembly. Reactions summarized in this schematic are, from left to right, nucleotide exchange in soluble FtsZ, magnesium-linked oligomerization in its GDP state, formation of FtsZ filaments mediated by GTP, and assembly into bundles in the presence of crowders. For simplicity, the soluble FtsZ species are represented as single beads. The plot of the bundle formation step shows the dependence of the fractional abundance of polymer bundles (1 g/l FtsZ) on Ficoll 70 concentration, as measured by low speed centrifugation b, c. AFM images of FtsZ polymers in the absence and presence, respectively, of 200 g/l Ficoll 70. d, e. EM images of FtsZ polymers in the absence and presence, respectively, of 220 g/l Ficoll 70. Bars: 100 nm (b–d) and 260 nm (e). Adapted from González et al. (2003, 2005) and Monterroso et al. (2012b)
Studying FtsZ in membrane-like systems
As the association of the proto-ring elements to form the functional complex in vivo takes place at the inner membrane, the interactions and assembly properties of FtsZ are currently being studied under topologically restricted reconstructions of the proto-ring structured in membrane-like systems, such as nanodiscs, coated beads, bilayers, and vesicles.
Biochemical analysis of proto-ring elements in nanodiscs and membrane-coated microbeads
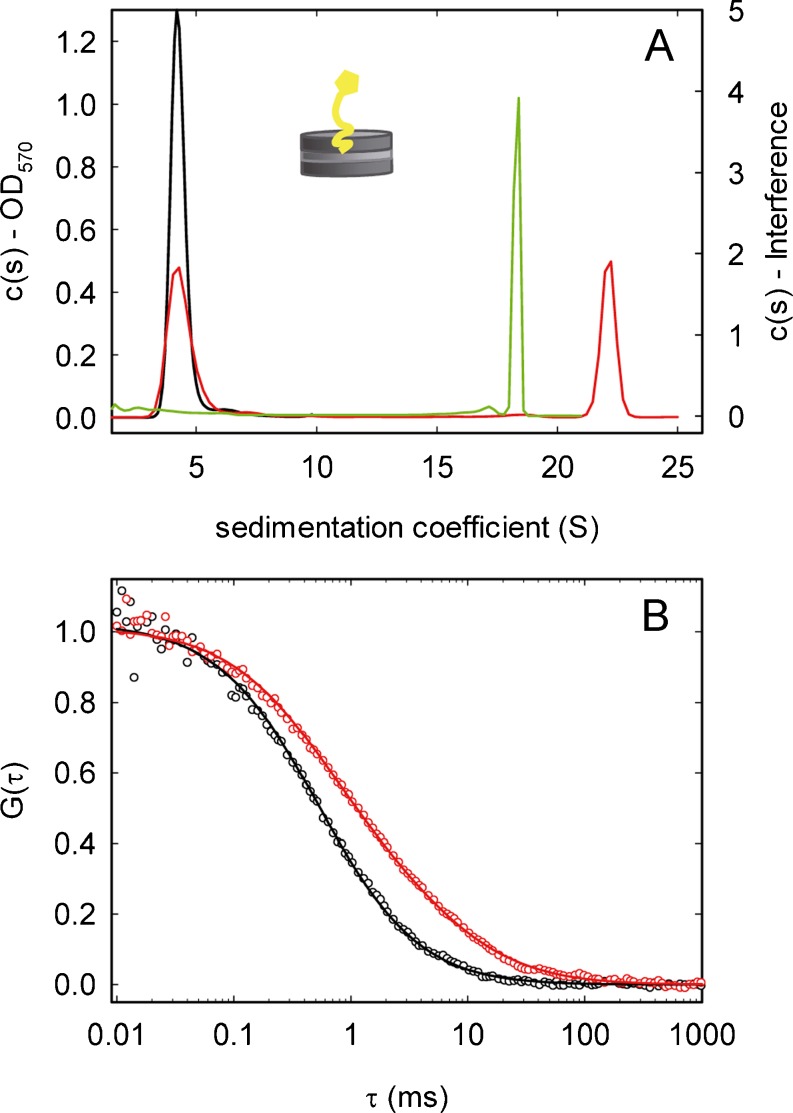
Nanodiscs consist of a ring formed by two copies of a membrane scaffold protein that encircles a phospholipid mixture, which can incorporate membrane proteins preserving their natural properties (Bayburt and Sligar 2010; Nath et al. 2007). They behave as soluble structures, allowing the quantitative analysis of their properties and interactions with other division elements in lipid environments by biochemical and biophysical techniques. A single copy of full-length ZipA was integrated in nanodiscs made of E. coli lipids (Nd-ZipA) as measured by analytical ultracentrifugation. This finding was confirmed by EM and is compatible with fluorescence correlation spectroscopy measurements of the diffusion coefficient of ZipA-containing nanodiscs (labeled with a trace amount of fluorescent lipid) (Hernández-Rocamora et al. 2012a, b) (Figs. 1, 6). These biophysical techniques were used to determine that ZipA embedded in nanodiscs interacts equally with different forms of FtsZ assembled in the presence of GDP, GTP, or GTP analogs (Fig. 6). They also allowed verification that peptides comprising the FtsZ region of interaction with ZipA compete with FtsZ polymers for Nd-ZipA binding. The strength of the interactions between Nd-ZipA and FtsZ oligomers and polymers was found to be of the same moderate affinity as the binding of FtsZ to a soluble variant of ZipA lacking the trans-membrane region (Martos et al. 2010). These results suggest that neither the trans-membrane region of ZipA nor FtsZ polymerization have a significant influence on the binding of FtsZ to ZipA. Moreover, they also support the dynamic nature of the anchoring of FtsZ to the membrane, facilitating the space and time modulation of FtsZ distribution during the cell cycle and its subsequent relocation to midcell when required for the assembly of a division ring. These nanodisc systems have been used to generate a fluorescence-based assay to screen for inhibitors of ZipA–FtsZ complex formation, with potential application to other protein–protein interactions of the divisome.
Fig. 6.
Reconstruction of proto-ring elements in nanodiscs. a Sedimentation coefficient [c(s)] distributions of Nd*-ZipA in the absence (black) and presence (red) of 50 μM FtsZ–GMPCPP obtained from absorbance data. Green c(s) distribution of FtsZ–GMPCPP alone from interference data in the same experiment. OD Optical density. Inset Schematic representation of a nanodisc containing ZipA. b Normalized fluorescence correlation spectroscopy autocorrelation profiles of Nd*-ZipA in the absence (black) and presence (red) of 50 μM FtsZ–GMPCPP. Adapted from Hernández-Rocamora et al. (2012b)
Micron-size beads coated with natural membranes or artificial proteolipid bilayers provide a reliable, robust system to generate lipid surfaces with uniform curvatures, in which both the lipid composition and the material of the bead can be modified in a controlled manner to obtain the desired properties of the system (Arumugam et al. 2011; Liu and Fletcher 2009). Using these beads, it is possible to measure the interactions between soluble and membrane-bound proto-ring elements by, among others, differential centrifugation, as in the case of the binding of FtsA to beads coated with inner membranes or lipids (Martos et al. 2012b). These approaches are currently being applied to measure the binding of different forms of FtsZ to membrane structures. Finally, the interaction between FtsZ and inner membranes incorporated into silver-coated polystyrene microbeads was monitored by surface-enhanced Raman scattering sensing, a surface-sensitive technique that enhances Raman scattering by molecules adsorbed to metal surfaces (Ahijado-Guzman et al. 2012). This strategy enabled structural changes arising from the binding of FtsZ polymers and oligomers at the surface of the bead to be detected, and these structural changes turned out to be very different, a result that could be exploited to design rapid systematic assays to screen for inhibitors of division protein–protein interactions.
Structural studies of proto-ring elements on lipid bilayers
These membrane-like systems allow the application of surface-sensitive imaging techniques, such as AFM and total internal reflection fluorescence microscopy (TIRFM), for studying the structural organization and dynamics of proteins and lipids on the membrane, detecting as well changes upon the binding of soluble proteins or assemblies to the membrane surface (Sezgin and Schwille 2012). The interaction between soluble and membrane-bound proteins can also be measured by acoustic and plasmonic sensing assays (Galush et al. 2009; Herrig et al. 2006). FtsZ polymers anchored to lipid bilayers through a soluble variant of ZipA were found to form dynamic bidimensional networks which evolve and reorganize with time by fragmentation, annealing and lateral condensation, as revealed by AFM (Mateos-Gil et al. 2012). Co-polymers of wild-type FtsZ and a variant form containing a membrane targeting sequence were found to present an intrinsic curvature as their alignment on microstructured substrates supporting the lipid bilayers, visualized by AFM and TIRFM, depends largely on the curvature of the surface. They preferentially align themselves along curvatures that reproduce the ones found in the inner face of E. coli cells (Arumugam et al. 2012).
Proto-ring elements in vesicles
The first evidence supporting the role of FtsZ as a contractive element in a lipid vesicle was provided by optical micrographs showing that polymers of an artificially membrane-attached variant of FtsZ (FtsZ-mts) were able to narrow the regions of elongated liposomes in which the FtsZ variant was located (Osawa et al. 2008). The cylindrical geometry of the tubular liposomes was found to be important as FtsZ rings were not observed inside spherical liposomes. The incorporation of these polymers on the external face of the liposomes caused their deformation, and lipid tubules were found (Osawa and Erickson 2011), a result also observed in the outside of vesicles with the natural forms of FtsZ and ZipA (Fig. 1). The geometry of the distortion depends upon the FtsZ terminal region at which the membrane targeting sequence is attached, indicating that FtsZ polymer bending (associated to the natural curvature of FtsZ) is an important factor in membrane deformation.
Giant vesicles, due to their large size (from several to hundreds of micrometers) are very suitable for investigating, by imaging and microspectroscopy techniques, the spatial/dynamic organization of proto-ring proteins trapped in their interior, their interacting properties and the changes in the distribution of membrane components linked to binding events, among others (Sezgin and Schwille 2012; Walde et al. 2010). The native form of FtsZ has been incorporated, together with FtsA, inside electroformed giant vesicles obtained from inner bacterial membrane, which naturally contains ZipA (Jiménez et al. 2011) (Fig. 1). The assembly of FtsZ modified the spatial distribution of the soluble proto-ring elements, resulting in the displacement of FtsA from the membrane, which was to be found associated with FtsZ polymers at the vesicle lumen, as observed by confocal microscopy. This result suggests that FtsA may have roles in division in addition to the known one in the association of FtsZ to the membrane during cytokinesis; as FtsA is an amphitropic protein, both FtsA and FtsZ may be related with signaling/activation events during the division process. On the other hand, membrane dilation of ZipA-containing vesicles was observed upon FtsZ polymerization mediated by caged-GTP inside the vesicle, suggesting a possible role for FtsZ in the modulation of membrane plasticity (Lopez-Montero et al. 2013).
Future challenges—towards reconstructing minimal division assemblies in cell-like (cytomimetic) compartments
The synthetic approaches described above have been exploited to study the properties of proto-ring elements and their interactions in membrane environments, allowing new insights to be gained into their precise mechanism of action and cellular function. However, many more experimental challenges have to be overcome before functional division machineries—divisomes—can be successfully constructed in the test tube.
-
The assembly of the divisome components in the bacteria occurs at discrete regions along the cell length (usually at midcell in normal E. coli division but it might vary in mutant cells). Therefore, strategies to reproduce these essential features in minimal reconstructions will require the construction of controllable compartments in which the molecular machinery that initiates division (the proto-ring) together with complexes that select the constriction site in vivo, namely the Min and nucleoid occlusion systems, will be assembled and their functions tested. Along these lines, Min waves, reproducing the oscillating behavior linked to the regulation of septum placement mediated by the Min system, have been partly reconstructed on the surface of lipid bilayers (Loose et al. 2008, 2011a, b) and, more recently, in microengineered lipid compartments of different shapes (Schweizer et al. 2012; Zieske and Schwille 2012). These oscillations have not yet been reproduced inside giant vesicles.
As mentioned above, the nucleoid is a large and highly charged entity that, because it occupies a significant volume of the intracellular space, may have a considerable influence on the spatial distribution of protein networks in the cytoplasm and at the membrane (Zimmerman 2006). Therefore, an important step will be to design a-cellular in vitro systems that mimic the spatial organization of proto-ring elements linked to volume exclusion and electrostatic effects in the crowded cytoplasm. Research towards a better understanding on how to best reproduce the crowded intracellular environment, including the production of artificial organelles (Marguet et al. 2012; Peters et al. 2012; Stadler et al. 2009) and micro/nanofabricated environments (Collier and Simpson 2011), is required to establish the influence of the local microenvironments at the bacterial interior (characterized by crowding and surface effects, small volumes, low copy numbers, anisotropic molecular distribution, phase transitions, microcompartments) on the structural, dynamic, and spatial organization of divisome assemblies. Systematic quantitative studies on the properties of highly concentrated heterogeneous mixtures of proteins and/or other macromolecules (Zhou et al. 2008) and on the organization, in time and space, of intracellular entities mediated by phase separation processes and the collective action of weak macromolecular interactions in crowded environments (Hyman and Simons 2012; Keating 2012) will be highly relevant.
-
Advances towards the optimization of procedures to encapsulate, in a controlled manner and with high yield, protein complexes inside cell-like compartments will be crucial (Abkarian et al. 2011; Matosevic 2012; Richmond et al. 2011; Takiguchi et al. 2011). Techniques to produce permeable giant vesicles are currently being explored to allow the control of FtsZ polymerization inside them by externally added ligands that promote assembly, such as GTP and Mg2+. Additionally, the application of microfabrication and patterning tools (Collier and Simpson 2011) combined with lipid chemistry technology will be highly relevant to our understanding of the influence of the size and shape of the cell-like compartments on the behavior of divisome assemblies. It has recently been shown that Min waves and patterns are very dependent on the structure, shape, and dimensions of the lipid containers (Schweizer et al. 2012; Zieske and Schwille 2012).
An alternative tool to produce minimal reconstructions of the divisome is the water-in-oil formation of microdroplets in microfluidic devices, resulting in droplets of sizes similar to giant vesicles, but completely monodisperse and easily manipulated inside microfluidic channels. These form picoliter compartments of which the composition and size can thus be controlled precisely, while fluorescence microscopy techniques offer systematic quantitative assays of individual droplets (Theberge et al. 2010). Related developments in droplet interface bilayer (Bayley et al. 2008) and polymersome (Martino et al. 2012) technologies also provide very interesting platforms for reconstruction purposes.
Finally, the positioning of the division site by the action of the Min protein system and/or the nucleoid will remain to be reproduced by introducing the proto-ring elements and positioning regulators into deformable closed compartments. These goals will require, in the case of the Min system, identifying the optimal physical conditions to convert traveling Min waves into steady oscillations.
A deeper quantitative understanding of the reversible association reactions involved in proto-ring assembly and its modulation by companion proteins and membrane elements is an additional limiting factor for future progress. This is not a trivial task, given the highly dynamic nature of most of these interactions, with a coexistence of a multiplicity of association states of which the relative abundance is composition and time-dependent. Such a rheostat-like behavior (in contrast with a switch-like engine) may facilitate function-linked structural rearrangements of division complexes during the cell cycle. A variety of new and improved biophysical tools exist that are suitable for achieving these goals, such as super-resolution and single molecule imaging (van Oijen 2010), fluorescence-based assays (Cisse et al. 2007), and plasmonic sensing (Baciu et al. 2008), among others.
The function of the force-generating elements driving constriction of the membrane needs to be reproduced in future minimal reconstructions of the divisome. The aims of current research is to complete a model of FtsZ action that integrates its mechanical properties in vitro with its role in division and with the constriction forces driving cytokinesis (Erickson 2009). The potential impact of other divisome proteins and, mainly, membrane elements on the forces exerted by FtsZ both in vitro and in the cell will have to be experimentally addressed. The lipid composition of the bacterial membrane can modulate its mechanical properties and also generate domains to recruit specific proteins (Janmey and Kinnunen 2006). Interestingly, it has been shown that FtsZ polymers modify the viscoelastic properties of E. coli lipid monolayers so that they become more fluid and plastic (Lopez-Montero et al. 2010, 2012). The very high content of cytoplasmic membrane proteins is also worth mentioning (Devaux and Seigneuret 1985) as this will very likely have a major effect on the mechanical properties of the membrane. Finally, the mechanical role of cell-wall synthesis during septum formation is poorly understood. Along these lines, the construction of fortifying shells around the membranes with the aim of reproducing the peptidoglycan layer seems to be an attractive but difficult approach to our better understanding of the mechanical processes linked to bacterial division.
In addition, other synthetic approaches to reconstruct minimal divisomes in cell-like compartments will come from the use of either cell-free expression systems inside giant vesicles (Forlin et al. 2012; Maeda et al. 2012; Noireaux et al. 2011) or cells in which the chromosome has been removed (Pazos et al. 2012). All of these developments, together with the application of powerful super-resolution microscopy and single molecule imaging tools inside the cell, will help to provide a detailed molecular description of essential bacterial division events.
Concluding remarks
Bacterial life is characterized by orderly patterns associated to processes carried out by macromolecular assemblies and networks, the components of which transiently interact to maintain, in time and space, the structural organization required for the optimal functioning of the cell (Kuthan 2001). The division of E. coli is a good example of such orderly processes, in which several proteins interact at the end of the cell cycle to form at midcell (with the cooperation of two positioning mechanisms) a dynamic ring which, in subsequent stages, recruits other proteins, resulting in the assembly of the molecular machinery effecting cytokinesis. Division-related molecular interactions take place in environments characterized by the presence of high concentrations of background molecules, structured as dynamic networks, and by the presence of surfaces and membranes. These microenvironments may influence the reactivity and location of proteins that are essential for septation, thus acting as nonspecific modulating factors of the division process.
An integrative approach towards elaborating the detailed biochemical mechanisms of bacterial division reactions that emerges from the study of essential proteins and functional dynamic assemblies in cell-like compartments is proposed. This approach attempts to take into account the relationship between individual protein species (and transient protein networks) and their local environment within the intact cell. This strategy, which enables complex processes to be investigated in facsimile cell medium whose composition may be manipulated in a controlled fashion, will help to reproduce the central features of bacterial division in the absence of cells.
Acknowledgments
GR will always be in debt to Allen Minton for his teachings and advice on many aspects of the scientific process, for his motivation and enthusiasm about science and life in general, and for being so generous with his time and knowledge. It is a real privilege to have such an exceptional maestro, a genuine scholar. Thanks for our friendship. The other authors of this review, particularly BM, share these feelings and are deeply grateful to Allen. His frequent visits to our lab have always been very inspirational and educational for all of us.
We also thank the members of our laboratory contributing to the works here reviewed, and Miguel Vicente (CNB-CSIC) for useful discussions and advice on bacterial division. This work was supported by the Spanish government through grants BIO2008-04478-C03 and BIO2011-28941-C03-03 to GR, and BFU2010-14910 and BIO2011-28941-C03-02 to SZ; by the European Commission through contract HEALTH-F3-2009-223432, by the Human Frontiers Science Program through grant RGP0050/2010-C102, and by Comunidad de Madrid through grant S-BIO-0260/2006 to GR; by the CSIC through grants 200980I186 and 201020I001 to SZ and CA, respectively. BM is a JAE postdoctoral associate from the European Social Fund and the Spanish Consejo Superior de Investigaciones Científicas (CSIC).
Conflicts of interest
None.
Footnotes
Special Issue: Protein-Protein and Protein-Ligand Interactions in Dilute and Crowded Solution Conditions. In Honor of Allen Minton’s 70th Birthday.
References
- Abkarian M, Loiseau E, Massiera G. Continuous droplet interface crossing encapsulation (cDICE) for high throughput monodisperse vesicle design. Soft Matter. 2011;7:4610–4614. doi: 10.1039/c1sm05239j. [DOI] [Google Scholar]
- Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7(9):642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- Ahijado-Guzman R, Gomez-Puertas P, Alvarez-Puebla RA, Rivas G, Liz-Marzan LM. Surface-enhanced raman scattering-based detection of the interactions between the essential cell division ftsz protein and bacterial membrane elements. ACS Nano. 2012;6(8):7514–7520. doi: 10.1021/nn302825u. [DOI] [PubMed] [Google Scholar]
- Alcorlo M, Jimenez M, Ortega A, Hermoso JM, Salas M, Minton AP, Rivas G. Analytical ultracentrifugation studies of phage phi29 protein p6 binding to DNA. J Mol Biol. 2009;385(5):1616–1629. doi: 10.1016/j.jmb.2008.11.044. [DOI] [PubMed] [Google Scholar]
- Arumugam S, Chwastek G, Schwille P. Protein-membrane interactions: the virtue of minimal systems in systems biology. Wiley Interdiscip Rev Syst Biol Med. 2011;3(3):269–280. doi: 10.1002/wsbm.119. [DOI] [PubMed] [Google Scholar]
- Arumugam S, Chwastek G, Fischer-Friedrich E, Ehrig C, Monch I, Schwille P. Surface topology engineering of membranes for the mechanical investigation of the tubulin homologue FtsZ. Angew Chem. 2012;51(47):11858–11862. doi: 10.1002/anie.201204332. [DOI] [PubMed] [Google Scholar]
- Attri AK, Minton AP. Composition gradient static light scattering: a new technique for rapid detection and quantitative characterization of reversible macromolecular hetero-associations in solution. Anal Biochem. 2005;346(1):132–138. doi: 10.1016/j.ab.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Attri AK, Minton AP. New methods for measuring macromolecular interactions in solution via static light scattering: basic methodology and application to nonassociating and self-associating proteins. Anal Biochem. 2005;337(1):103–110. doi: 10.1016/j.ab.2004.09.045. [DOI] [PubMed] [Google Scholar]
- Baciu CL, Becker J, Janshoff A, Sonnichsen C. Protein-membrane interaction probed by single plasmonic nanoparticles. Nano Lett. 2008;8(6):1724–1728. doi: 10.1021/nl080805l. [DOI] [PubMed] [Google Scholar]
- Bayburt TH, Sligar SG. Membrane protein assembly into nanodiscs. FEBS Lett. 2010;584(9):1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley H, Cronin B, Heron A, Holden MA, Hwang WL, Syeda R, Thompson J, Wallace M. Droplet interface bilayers. Mol Biosyst. 2008;4(12):1191–1208. doi: 10.1039/b808893d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YY, Cronan JE., Jr Molecular cloning, DNA sequencing, and enzymatic analyses of two Escherichia coli pyruvate oxidase mutants defective in activation by lipids. J Bacteriol. 1986;167(1):312–318. doi: 10.1128/jb.167.1.312-318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Erickson HP. Rapid in vitro assembly dynamics and subunit turnover of FtsZ demonstrated by fluorescence resonance energy transfer. J Biol Chem. 2005;280(23):22549–22554. doi: 10.1074/jbc.M500895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse I, Okumus B, Joo C, Ha T. Fueling protein DNA interactions inside porous nanocontainers. Proc Natl Acad Sci USA. 2007;104(31):12646–12650. doi: 10.1073/pnas.0610673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier CP, Simpson ML. Micro/nanofabricated environments for synthetic biology. Curr Opin Biotechnol. 2011;22(4):516–526. doi: 10.1016/j.copbio.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Devaux PF, Seigneuret M. Specificity of lipid-protein interactions as determined by spectroscopic techniques. Biochim Biophys Acta. 1985;822(1):63–125. doi: 10.1016/0304-4157(85)90004-8. [DOI] [PubMed] [Google Scholar]
- Dix JA, Hom EFY, Verkman AS. Fluorescence correlation spectroscopy simulations of photophysical phenomena and molecular interactions: a molecular dynamics/Monte Carlo approach. J Phys Chem B. 2006;110(4):1896–1906. doi: 10.1021/jp055840k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan AJ, Vollmer W. The physiology of bacterial cell division. Ann N Y Acad Sci. 2013;1277(1):8–28. doi: 10.1111/j.1749-6632.2012.06818.x. [DOI] [PubMed] [Google Scholar]
- Elcock AH. Models of macromolecular crowding effects and the need for quantitative comparisons with experiment. Curr Opin Struct Biol. 2010;20(2):196–206. doi: 10.1016/j.sbi.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci. 2001;26(10):597–604. doi: 10.1016/S0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- Erickson HP. Modeling the physics of FtsZ assembly and force generation. Proc Natl Acad Sci USA. 2009;106(23):9238–9243. doi: 10.1073/pnas.0902258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev. 2010;74(4):504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodeke AA, Minton AP. Quantitative characterization of polymer-polymer, protein-protein, and polymer-protein interaction via tracer sedimentation equilibrium. J Phys Chem B. 2010;114(33):10876–10880. doi: 10.1021/jp104342f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forlin M, Lentini R, Mansy SS. Cellular imitations. Curr Opin Chem Biol. 2012;16(5–6):586–592. doi: 10.1016/j.cbpa.2012.10.020. [DOI] [PubMed] [Google Scholar]
- Galush WJ, Shelby SA, Mulvihill MJ, Tao A, Yang P, Groves JT. A nanocube plasmonic sensor for molecular binding on membrane surfaces. Nano Lett. 2009;9(5):2077–2082. doi: 10.1021/nl900513k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler B, Shiomi D, Margolin W. The ftsA* gain-of-function allele of Escherichia coli and its effects on the stability and dynamics of the Z ring. Microbiology. 2007;153(Pt 3):814–825. doi: 10.1099/mic.0.2006/001834-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González JM, Jiménez M, Vélez M, Mingorance J, Andreu JM, Vicente M, Rivas G. Essential cell division protein FtsZ assembles into one monomer-thick ribbons under conditions resembling the crowded intracellular environment. J Biol Chem. 2003;278(39):37664–37671. doi: 10.1074/jbc.M305230200. [DOI] [PubMed] [Google Scholar]
- González JM, Velez M, Jiménez M, Alfonso C, Schuck P, Mingorance J, Vicente M, Minton AP, Rivas G. Cooperative behavior of Escherichia coli cell-division protein FtsZ assembly involves the preferential cyclization of long single-stranded fibrils. Proc Natl Acad Sci USA. 2005;102(6):1895–1900. doi: 10.1073/pnas.0409517102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell DS. The machinery of life. 2. New York: Copernicus Books (Springer Science + Business Media); 2010. [Google Scholar]
- Hall D, Minton AP. Macromolecular crowding: qualitative and semiquantitative successes, quantitative challenges. Biochim Biophys Acta. 2003;1649(2):127–139. doi: 10.1016/S1570-9639(03)00167-5. [DOI] [PubMed] [Google Scholar]
- Haney SA, Glasfeld E, Hale C, Keeney D, He Z, de Boer P. Genetic analysis of the Escherichia coli FtsZ.ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J Biol Chem. 2001;276(15):11980–11987. doi: 10.1074/jbc.M009810200. [DOI] [PubMed] [Google Scholar]
- Hernández-Rocamora VM, García-Montañés C, Rivas G, Llorca O. Reconstitution of the Escherichia coli cell division ZipA-FtsZ complexes in nanodiscs as revealed by electron microscopy. J Struct Biol. 2012;180(3):531–538. doi: 10.1016/j.jsb.2012.08.013. [DOI] [PubMed] [Google Scholar]
- Hernández-Rocamora VM, Reija B, García C, Natale P, Alfonso C, Minton AP, Zorrilla S, Rivas G, Vicente M. Dynamic interaction of the Escherichia coli cell division ZipA and FtsZ proteins evidenced in nanodiscs. J Biol Chem. 2012;287(36):30097–30104. doi: 10.1074/jbc.M112.388959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrig A, Janke M, Austermann J, Gerke V, Janshoff A, Steinem C. Cooperative adsorption of ezrin on PIP2-containing membranes. Biochemistry. 2006;45(43):13025–13034. doi: 10.1021/bi061064a. [DOI] [PubMed] [Google Scholar]
- Herzfeld J. Crowding-induced organization in cells: spontaneous alignment and sorting of filaments with physiological control points. J Mol Recognit. 2004;17(5):376–381. doi: 10.1002/jmr.703. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Simons K. Cell biology. Beyond oil and water–phase transitions in cells. Science. 2012;337(6098):1047–1049. doi: 10.1126/science.1223728. [DOI] [PubMed] [Google Scholar]
- Janmey PA, Kinnunen PK. Biophysical properties of lipids and dynamic membranes. Trends Cell Biol. 2006;16(10):538–546. doi: 10.1016/j.tcb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Jiao M, Li HT, Chen J, Minton AP, Liang Y. Attractive protein-polymer interactions markedly alter the effect of macromolecular crowding on protein association equilibria. Biophys J. 2010;99(3):914–923. doi: 10.1016/j.bpj.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez M, Rivas G, Minton AP. Quantitative characterization of weak self-association in concentrated solutions of immunoglobulin G via the measurement of sedimentation equilibrium and osmotic pressure. Biochemistry. 2007;46(28):8373–8378. doi: 10.1021/bi7005515. [DOI] [PubMed] [Google Scholar]
- Jiménez M, Martos A, Vicente M, Rivas G. Reconstitution and organization of Escherichia coli proto-ring elements (FtsZ and FtsA) inside giant unilamellar vesicles obtained from bacterial inner membranes. J Biol Chem. 2011;286(13):11236–11241. doi: 10.1074/jbc.M110.194365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating CD. Aqueous phase separation as a possible route to compartmentalization of biological molecules. Acc Chem Res. 2012;45(12):2114–2124. doi: 10.1021/ar200294y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupka M, Rivas G, Rico AI, Vicente M. Key role of two terminal domains in the bidirectional polymerization of FtsA protein. J Biol Chem. 2012;287(10):7756–7765. doi: 10.1074/jbc.M111.311563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuthan H. Self-organisation and orderly processes by individual protein complexes in the bacterial cell. Prog Biophys Mol Biol. 2001;75(1–2):1–17. doi: 10.1016/S0079-6107(00)00023-7. [DOI] [PubMed] [Google Scholar]
- Lavalette D, Hink MA, Tourbez M, Tetreau C, Visser AJ. Proteins as micro viscosimeters: brownian motion revisited. Eur Biophys J Biophys Lett. 2006;35(6):517–522. doi: 10.1007/s00249-006-0060-z. [DOI] [PubMed] [Google Scholar]
- Liu AP, Fletcher DA. Biology under construction: in vitro reconstitution of cellular function. Nat Rev. 2009;10(9):644–650. doi: 10.1038/nrm2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science (New York) 2008;320(5877):789–792. doi: 10.1126/science.1154413. [DOI] [PubMed] [Google Scholar]
- Loose M, Fischer-Friedrich E, Herold C, Kruse K, Schwille P. Min protein patterns emerge from rapid rebinding and membrane interaction of MinE. Nat Struct Mol Biol. 2011;18(5):577–583. doi: 10.1038/nsmb.2037. [DOI] [PubMed] [Google Scholar]
- Loose M, Kruse K, Schwille P. Protein self-organization: lessons from the min system. Annu Rev Biophys. 2011;40:315–336. doi: 10.1146/annurev-biophys-042910-155332. [DOI] [PubMed] [Google Scholar]
- Lopez-Montero I, Arriaga LR, Rivas G, Velez M, Monroy F. Lipid domains and mechanical plasticity of Escherichia coli lipid monolayers. Chem Phys Lipids. 2010;163(1):56–63. doi: 10.1016/j.chemphyslip.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Lopez-Montero I, Mateos-Gil P, Sferrazza M, Navajas PL, Rivas G, Velez M, Monroy F. Active membrane viscoelasticity by the bacterial FtsZ-division protein. Langmuir. 2012;28(10):4744–4753. doi: 10.1021/la204742b. [DOI] [PubMed] [Google Scholar]
- Lopez-Montero I, Lopez-Navajas P, Mingorance J, Velez M, Vicente M, Monroy F. Membrane reconstruction of FtsZ-ZipA complex inside giant spherical vesicles made of E. coli lipid: large membrane dilation and analysis of membrane plasticity. Biochim Biophys Acta. 2013;1828:687–698. doi: 10.1016/j.bbamem.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Lu C, Stricker J, Erickson HP. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima—quantitation, GTP hydrolysis, and assembly. Cell Motil Cytoskeleton. 1998;40(1):71–86. doi: 10.1002/(SICI)1097-0169(1998)40:1<71::AID-CM7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J, Pichoff S, Du S. Bacterial cytokinesis: from Z ring to divisome. Cytoskeleton (Hoboken) 2012;69(10):778–790. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda YT, Nakadai T, Shin J, Uryu K, Noireaux V, Libchaber A. Assembly of MreB filaments on liposome membranes: a synthetic biology approach. ACS Synth Biol. 2012;1(2):53–59. doi: 10.1021/sb200003v. [DOI] [PubMed] [Google Scholar]
- Margolin W. Organelle division: self-assembling GTPase caught in the middle. Curr Biol. 2000;10(9):R328–R330. doi: 10.1016/S0960-9822(00)00458-9. [DOI] [PubMed] [Google Scholar]
- Marguet M, Sandre O, Lecommandoux S. Polymersomes in “gelly” polymersomes: toward structural cell mimicry. Langmuir. 2012;28(4):2035–2043. doi: 10.1021/la204018w. [DOI] [PubMed] [Google Scholar]
- Martino C, Kim SH, Horsfall L, Abbaspourrad A, Rosser SJ, Cooper J, Weitz DA. Protein expression, aggregation, and triggered release from polymersomes as artificial cell-like structures. Angew Chem Int Ed Engl. 2012;51:6416–6420. doi: 10.1002/anie.201201443. [DOI] [PubMed] [Google Scholar]
- Martos A, Alfonso C, López-Navajas P, Ahijado-Guzman R, Mingorance J, Minton AP, Rivas G. Characterization of self-association and heteroassociation of bacterial cell division proteins FtsZ and ZipA in solution by composition gradient-static light scattering. Biochemistry. 2010;49(51):10780–10787. doi: 10.1021/bi101495x. [DOI] [PubMed] [Google Scholar]
- Martos A, Jimenez M, Rivas G, Schwille P. Towards a bottom-up reconstitution of bacterial cell division. Trends Cell Biol. 2012;22(12):634–643. doi: 10.1016/j.tcb.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Martos A, Monterroso B, Zorrilla S, Reija B, Alfonso C, Mingorance J, Rivas G, Jimenez M. Isolation, characterization and lipid-binding properties of the recalcitrant FtsA division protein from Escherichia coli. PLoS One. 2012;7(6):e39829. doi: 10.1371/journal.pone.0039829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Gil P, Paez A, Horger I, Rivas G, Vicente M, Tarazona P, Velez M. Depolymerization dynamics of individual filaments of bacterial cytoskeletal protein FtsZ. Proc Natl Acad Sci USA. 2012;109:8133–8138. doi: 10.1073/pnas.1204844109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matosevic S. Synthesizing artificial cells from giant unilamellar vesicles: state-of-the art in the development of microfluidic technology. Bioessays. 2012;34(11):992–1001. doi: 10.1002/bies.201200105. [DOI] [PubMed] [Google Scholar]
- Menendez M, Rivas G, Diaz JF, Andreu JM. Control of the structural stability of the tubulin dimer by one high affinity bound magnesium ion at nucleotide N-site. J Biol Chem. 1998;273(1):167–176. doi: 10.1074/jbc.273.1.167. [DOI] [PubMed] [Google Scholar]
- Mingorance J, Rivas G, Velez M, Gomez-Puertas P, Vicente M. Strong FtsZ is with the force: mechanisms to constrict bacteria. Trends Microbiol. 2010;18(8):348–356. doi: 10.1016/j.tim.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Minton AP. The effect of volume occupancy upon the thermodynamic activity of proteins: some biochemical consequences. Mol Cell Biochem. 1983;55(2):119–140. doi: 10.1007/BF00673707. [DOI] [PubMed] [Google Scholar]
- Minton AP. Holobiochemistry: the effect of local environment upon the equilibria and rates of biochemical reactions. Int J Biochem. 1990;22(10):1063–1067. doi: 10.1016/0020-711X(90)90102-9. [DOI] [PubMed] [Google Scholar]
- Minton AP. Implications of macromolecular crowding for protein assembly. Curr Opin Struct Biol. 2000;10(1):34–39. doi: 10.1016/S0959-440X(99)00045-7. [DOI] [PubMed] [Google Scholar]
- Minton AP. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J Biol Chem. 2001;276(14):10577–10580. doi: 10.1074/jbc.R100005200. [DOI] [PubMed] [Google Scholar]
- Minton AP. How can biochemical reactions within cells differ from those in test tubes? J Cell Sci. 2006;119(Pt 14):2863–2869. doi: 10.1242/jcs.03063. [DOI] [PubMed] [Google Scholar]
- Minton AP. Hard quasispherical particle models for the viscosity of solutions of protein mixtures. J Phys Chem B. 2012;116(31):9310–9315. doi: 10.1021/jp302748k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton AP. Quantitative assessment of the relative contributions of steric repulsion and chemical interactions to macromolecular crowding. Biopolymers. 2013;99(4):239–244. doi: 10.1002/bip.22163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton AP, Wilf J. Effect of macromolecular crowding upon the structure and function of an enzyme: glyceraldehyde-3-phosphate dehydrogenase. Biochemistry. 1981;20(17):4821–4826. doi: 10.1021/bi00520a003. [DOI] [PubMed] [Google Scholar]
- Monterroso B, Ahijado-Guzman R, Reija B, Alfonso C, Zorrilla S, Minton AP, Rivas G. Mg(2+)-linked self-assembly of FtsZ in the presence of GTP or a GTP analogue involves the concerted formation of a narrow size distribution of oligomeric species. Biochemistry. 2012;51:4541–4550. doi: 10.1021/bi300401b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monterroso B, Rivas G, Minton AP. An equilibrium model for the Mg(2+)-linked self-assembly of FtsZ in the presence of GTP or a GTP analogue. Biochemistry. 2012;51:6108–6113. doi: 10.1021/bi300891q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan S, Amir D, Grupi A, Goldenberg DP, Minton AP, Haas E. Modulation of functionally significant conformational equilibria in adenylate kinase by high concentrations of trimethylamine oxide attributed to volume exclusion. Biophys J. 2011;100(12):2991–2999. doi: 10.1016/j.bpj.2011.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanninga N. Morphogenesis of Escherichia coli. Microbiol Mol Biol Rev. 1998;62(1):110–129. doi: 10.1128/mmbr.62.1.110-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath A, Atkins WM, Sligar SG. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46(8):2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- Noireaux V, Maeda YT, Libchaber A. Development of an artificial cell, from self-organization to computation and self-reproduction. Proc Natl Acad Sci USA. 2011;108(9):3473–3480. doi: 10.1073/pnas.1017075108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi T, Hale CA, de Boer PA, Erickson HP. Structural evidence that the P/Q domain of ZipA is an unstructured, flexible tether between the membrane and the C-terminal FtsZ-binding domain. J Bacteriol. 2002;184(15):4313–4315. doi: 10.1128/JB.184.15.4313-4315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Erickson HP. Inside-out Z rings–constriction with and without GTP hydrolysis. Mol Microbiol. 2011;81(2):571–579. doi: 10.1111/j.1365-2958.2011.07716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ Rings in Liposomes. Science. 2008;320(5877):792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos M, Natale P, Vicente M. A specific role for the ZipA protein in cell division: stabilization of the FtsZ protein. J Biol Chem. 2012;288(5):3219–3226. doi: 10.1074/jbc.M112.434944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters RJRW, Louzao I, van Hest JCM. From polymeric nanoreactors to artificial organelles. Chem Sci. 2012;3(2):335–342. doi: 10.1039/c2sc00803c. [DOI] [Google Scholar]
- Phillip Y, Schreiber G. Formation of protein complexes in crowded environments—From in vitro to in vivo. FEBS Lett. 2013 doi: 10.1016/j.febslet.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol. 2005;55(6):1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- Pichoff S, Shen B, Sullivan B, Lutkenhaus J. FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA’s self-interaction competes with its ability to recruit downstream division proteins. Mol Microbiol. 2012;83(1):151–167. doi: 10.1111/j.1365-2958.2011.07923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp D, Iwasa M, Narita A, Erickson HP, Maeda Y. FtsZ condensates: an in vitro electron microscopy study. Biopolymers. 2009;91(5):340–350. doi: 10.1002/bip.21136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record MT, Courtenay ES, Cayley S, Guttman HJ. Biophysical compensation mechanisms buffering E-coli protein-nucleic acid interactions against changing environments. Trends Biochem Sci. 1998;23(5):190–194. doi: 10.1016/S0968-0004(98)01207-9. [DOI] [PubMed] [Google Scholar]
- Reija B, Monterroso B, Jiménez M, Vicente M, Rivas G, Zorrilla S. Development of a homogeneous fluorescence anisotropy assay to monitor and measure FtsZ assembly in solution. Anal Biochem. 2011;418(1):89–96. doi: 10.1016/j.ab.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Richmond DL, Schmid EM, Martens S, Stachowiak JC, Liska N, Fletcher DA. Forming giant vesicles with controlled membrane composition, asymmetry, and contents. Proc Natl Acad Sci USA. 2011;108(23):9431–9436. doi: 10.1073/pnas.1016410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas G, Minton AP. Non-ideal tracer sedimentation equilibrium: a powerful tool for the characterization of macromolecular interactions in crowded solutions. J Mol Recognit. 2004;17(5):362–367. doi: 10.1002/jmr.708. [DOI] [PubMed] [Google Scholar]
- Rivas G, Fernandez JA, Minton AP. Direct observation of the self-association of dilute proteins in the presence of inert macromolecules at high concentration via tracer sedimentation equilibrium: theory, experiment, and biological significance. Biochemistry. 1999;38(29):9379–9388. doi: 10.1021/bi990355z. [DOI] [PubMed] [Google Scholar]
- Rivas G, López A, Mingorance J, Ferrándiz MJ, Zorrilla S, Minton AP, Vicente M, Andreu JM. Magnesium-induced linear self-association of the FtsZ bacterial cell division protein monomer. The primary steps for FtsZ assembly. The J Biol Chem. 2000;275(16):11740–11749. doi: 10.1074/jbc.275.16.11740. [DOI] [PubMed] [Google Scholar]
- Rivas G, Fernandez JA, Minton AP. Direct observation of the enhancement of noncooperative protein self-assembly by macromolecular crowding: Indefinite linear self-association of bacterial cell division protein FtsZ. Proc Natl Acad Sci USA. 2001;98(6):3150–3155. doi: 10.1073/pnas.051634398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda S, Vicente M, Mingorance J. Concentration and assembly of the division ring proteins FtsZ, FtsA, and ZipA during the Escherichia coli cell cycle. J Bacteriol. 2003;185(11):3344–3351. doi: 10.1128/JB.185.11.3344-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvarelli E, Krupka M, Rivas G, Vicente M, Mingorance J. Independence between GTPase active sites in the Escherichia coli cell division protein FtsZ. FEBS Lett. 2011;585(24):3880–3883. doi: 10.1016/j.febslet.2011.10.046. [DOI] [PubMed] [Google Scholar]
- Schweizer J, Loose M, Bonny M, Kruse K, Monch I, Schwille P. Geometry sensing by self-organized protein patterns. Proc Natl Acad Sci USA. 2012;109(38):15283–15288. doi: 10.1073/pnas.1206953109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezgin E, Schwille P. Model membrane platforms to study protein-membrane interactions. Mol Membr Biol. 2012;29(5):144–154. doi: 10.3109/09687688.2012.700490. [DOI] [PubMed] [Google Scholar]
- Spitzer J, Poolman B. The role of biomacromolecular crowding, ionic strength, and physicochemical gradients in the complexities of life’s emergence. Microbiol Mol Biol Rev. 2009;73(2):371–388. doi: 10.1128/MMBR.00010-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler B, Price AD, Chandrawati R, Hosta-Rigau L, Zelikin AN, Caruso F. Polymer hydrogel capsules: en route toward synthetic cellular systems. Nanoscale. 2009;1(1):68–73. doi: 10.1039/b9nr00143c. [DOI] [PubMed] [Google Scholar]
- Strauss MP, Liew AT, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ. 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biol. 2012;10(9):e1001389. doi: 10.1371/journal.pbio.1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker J, Maddox P, Salmon ED, Erickson HP. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acad Sci USA. 2002;99(5):3171–3175. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P, Wang Q, Freund SM, Lowe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31(10):2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takiguchi K, Negishi M, Tanaka-Takiguchi Y, Homma M, Yoshikawa K. Transformation of actoHMM assembly confined in cell-sized liposome. Langmuir. 2011;27(18):11528–11535. doi: 10.1021/la2016287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theberge AB, Courtois F, Schaerli Y, Fischlechner M, Abell C, Hollfelder F, Huck WT. Microdroplets in microfluidics: an evolving platform for discoveries in chemistry and biology. Angew Chem. 2010;49(34):5846–5868. doi: 10.1002/anie.200906653. [DOI] [PubMed] [Google Scholar]
- Tonthat NK, Arold ST, Pickering BF, Van Dyke MW, Liang S, Lu Y, Beuria TK, Margolin W, Schumacher MA. Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J. 2011;30(1):154–164. doi: 10.1038/emboj.2010.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oijen AM. Single-molecule approaches to characterizing kinetics of biomolecular interactions. Curr Opin Biotechnol. 2010;22(1):75–80. doi: 10.1016/j.copbio.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Vendeville A, Lariviere D, Fourmentin E. An inventory of the bacterial macromolecular components and their spatial organization. FEMS Microbiol Rev. 2011;35(2):395–414. doi: 10.1111/j.1574-6976.2010.00254.x. [DOI] [PubMed] [Google Scholar]
- Vicente M. Crystal ball: Will test tubes ever undergo division? Microbiol Biotechnol. 2013;6:3–16. doi: 10.1111/1751-7915.12014. [DOI] [Google Scholar]
- Vicente M, Rico AI. The order of the ring: assembly of Escherichia coli cell division components. Mol Microbiol. 2006;61(1):5–8. doi: 10.1111/j.1365-2958.2006.05233.x. [DOI] [PubMed] [Google Scholar]
- Walde P, Cosentino K, Engel H, Stano P. Giant vesicles: preparations and applications. ChemBioChem. 2010;11(7):848–865. doi: 10.1002/cbic.201000010. [DOI] [PubMed] [Google Scholar]
- Wilf J, Minton AP. Evidence for protein self-association induced by excluded volume. Myoglobin in the presence of globular proteins. Biochim Biophys Acta. 1981;670(3):316–322. doi: 10.1016/0005-2795(81)90103-3. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Nucleoid occlusion and bacterial cell division. Nat Rev Microbiol. 2012;10(1):8–12. doi: 10.1038/nrmicro2671. [DOI] [PubMed] [Google Scholar]
- Zhang B, Zhang Y, Wang Z, Zheng Y. The role of Mg2+ cofactor in the guanine nucleotide exchange and GTP hydrolysis reactions of Rho family GTP-binding proteins. J Biol Chem. 2000;275(33):25299–25307. doi: 10.1074/jbc.M001027200. [DOI] [PubMed] [Google Scholar]
- Zhou HX. Influence of crowded cellular environments on protein folding, binding, and oligomerization: biological consequences and potentials of atomistic modeling. FEBS Lett. 2013 doi: 10.1016/j.febslet.2013.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HX, Rivas G, Minton AP. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieske K, Schwille P. Reconstitution of pole-to-pole oscillations of min proteins in microengineered polydimethylsiloxane compartments. Angew Chem. 2012;52(1):459–462. doi: 10.1002/anie.201207078. [DOI] [PubMed] [Google Scholar]
- Zimmerman SB. Shape and compaction of Escherichia coli nucleoids. J Struct Biol. 2006;156(2):255–261. doi: 10.1016/j.jsb.2006.03.022. [DOI] [PubMed] [Google Scholar]
- Zorrilla S, Jimenez M, Lillo P, Rivas G, Minton AP. Sedimentation equilibrium in a solution containing an arbitrary number of solute species at arbitrary concentrations: theory and application to concentrated solutions of ribonuclease. Biophys Chem. 2004;108(1–3):89–100. doi: 10.1016/j.bpc.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Zorrilla S, Rivas G, Acuna AU, Lillo MP. Protein self-association in crowded protein solutions: a time-resolved fluorescence polarization study. Protein Sci. 2004;13(11):2960–2969. doi: 10.1110/ps.04809404. [DOI] [PMC free article] [PubMed] [Google Scholar]