Abstract
Background:
The SLUG transcription factor has been linked with the KIT signalling pathway that is important for gastrointestinal stromal tumour (GIST) tumourigenesis. Its clinical significance in GIST is unknown.
Methods:
Influence of SLUG expression on cell proliferation and viability were investigated in GIST48 and GIST882 cell lines. The association between tumour SLUG expression in immunohistochemistry and recurrence-free survival (RFS) was studied in two clinical GIST series, one with 187 patients treated with surgery alone, and another one with 313 patients treated with surgery and adjuvant imatinib.
Results:
SLUG downregulation inhibited cell proliferation, induced cell death in both cell lines, and sensitised GIST882 cells to lower imatinib concentrations. SLUG was expressed in 125 (25.0%) of the 500 clinical GISTs evaluated, and expression was associated with several factors linked with unfavourable prognosis. SLUG expression was associated with unfavourable RFS both when patients were treated with surgery alone (HR=3.40, 95% CI=1.67–6.89, P=0.001) and when treated with surgery plus adjuvant imatinib (HR=1.83, 95% CI=1.29–2.60, P=0.001).
Conclusions:
GIST patients with high tumour SLUG expression have unfavourable RFS. SLUG may mediate pro-survival signalling in GISTs.
Keywords: gastrointestinal stromal tumour, SLUG, SNAI2, survival, proliferation, apoptosis
Gastrointestinal stromal tumour (GIST) is one of the most common types of sarcoma (Ducimetiere et al, 2011). Approximately 90% of GISTs contain an activating mutation either in KIT or PDGFRA (platelet-derived growth factor receptor alpha) genes, which leads to ligand independent activation of these receptor tyrosine kinases (Hirota et al, 1998; Heinrich et al, 2003). GISTs that do not harbor a KIT or PDGFRA mutation may have mutation in other genes, such as succinate dehydrogenase (SDH) or neurofibromatosis type 1 (NF1; Nannini et al, 2014). Imatinib mesylate that targets KIT and PDGFRA is the standard first-line therapy for patients with metastatic GIST (Demetri et al, 2002), and the standard adjuvant therapy for those patients whose tumour is considered to have a high risk for recurrence after surgery and contains an imatinib-sensitive mutation (Dematteo et al, 2009; Joensuu et al, 2012a). However, drug resistance frequently emerges when patients with advanced GIST are treated with imatinib or other tyrosine kinase inhibitors (Heinrich et al, 2006).
SLUG (the protein product of SNAI2) is a member of the Snail family of zinc-finger transcription factors (Nieto, 2002). It is involved in the regulation of cell migration during embryogenesis and in tumour cell invasion and migration (Cobaleda et al, 2007). SNAIL (product of SNAI1) and SLUG are expressed in several types of human cancer, and their expression is often associated with low-histological grade of differentiation, tumour recurrence, metastasis and poor prognosis (Cobaleda et al, 2007; Wang et al, 2013). Although SNAIL and SLUG may induce the epithelial-mesenchymal transition, they have a role also in the maintenance of the stem cell-like properties of tumour cells. The stem cell-like phenotype of tumour cells has been linked with resistance to radiotherapy and chemotherapy in various types of cancer (Perez-Losada et al, 2003; Catalano et al, 2004; Kurrey et al, 2009; Zhou et al, 2014). Downregulation of SLUG inhibits proliferation of cell lines derived from colorectal cancer, prostate cancer and esophageal cancer, and, in accordance with this, overexpression of SLUG increases proliferation of human glioblastoma cells (Emadi Baygi et al, 2010; Yang et al, 2010; Zhang et al, 2011b; Qian et al, 2013). SLUG is involved in cell survival also through either direct or indirect transcriptional regulation of proapoptotic and antiapoptotic genes (Tribulo et al, 2004; Wu et al, 2005; Bermejo-Rodriguez et al, 2006; Vitali et al, 2008; Kim et al, 2014).
The stem cell factor (SCF, the ligand of KIT) and KIT signalling upregulate SLUG in malignant mesothelioma cells (Catalano et al, 2004), and a few other studies have suggested a relationship between SCF/KIT signalling and SLUG. Defects in SLUG and KIT signalling lead to phenotypes with similar features both in humans and in mice. Homozygous deletions of the SLUG gene in the human cause the Waardenburg disease, a rare disorder associated with abnormal pigmentation and sensorineural deafness (Sanchez-Martin et al, 2003). Similarly, dysfunctional KIT may cause piepaldism and sensorineural deafness (Spritz and Beighton, 1998). Mice with targeted SLUG deletions have phenotypic similarities with mice with defective KIT or SCF, including abnormal pigmentation, and gonadal and haematopoietic defects (Motro et al, 1991; Perez-Losada et al, 2002).
Because SLUG defects result in clinical phenotypes resembling those that result from KIT defects, and SLUG influences cancer cell proliferation and therapy resistance in various tumour types, we investigated whether SLUG influences also GIST cell proliferation and whether SLUG expression might be associated with the histopathological and clinical properties of GISTs and patient survival.
Materials and methods
Patients and tumour samples
The associations between GIST SLUG expression, tumour features and patient survival outcomes were investigated in two clinical series consisting of GIST patients. The first series is a population-based cohort consisting of patients who were treated with surgery in western Sweden in 1983–2000, as described in detail in (Nilsson et al, 2005a). Out of the total of 288 patients in the cohort, we included in the current study 187 (64.9%) patients who had tumour tissue available, tumour histology was compatible with GIST and stained positively for KIT in immunohistochemistry, and provided that the patients had information about gender, tumour diameter, and follow-up available, whereas the remaining 101 (35.1%) patients were excluded. None of the patients was treated with imatinib or other tyrosine kinase inhibitors after surgery for GIST. The median follow-up time of the patients was 44 months after surgery.
The second series consists of GIST patients who were entered to the Scandinavian Sarcoma Group (SSG) XVIII/Arbeitsgemeinschaft Internistische Onkologie (AIO) trial (Joensuu et al, 2012a, 2016). SSGXVIII/AIO is an open-label, multicenter, phase 3 study. Patients who had KIT-positive, operable tumour and who were at a high risk for recurrence according to the modified National Institute of Health (NIH) Criteria (Joensuu, 2008) were eligible. After macroscopically complete surgery the patients were randomly allocated to receive adjuvant imatinib 400 mg per day orally either for 12 or 36 months. A total of 400 patients from Finland, Germany, Norway and Sweden were entered to the trial between February 2004 and September 2008. Representative tumour tissue for the current study was available from 313 (85.1%) out of the 368 patients who provided consent, had KIT-positive GIST in central histopathology review of tumour tissue, and had localized disease at the time of randomisation. The median follow-up time after the date of randomisation was 6.1 years (Joensuu et al, 2016).
Institutional review committees of the Helsinki University Hospital, Helsinki, Finland, and Sahlgrenska University Hospital, Gothenburg, Sweden, approved the current study. The patients who participated in the SSGXVIII/AIO trial provided written informed consent for the study.
Immunohistochemistry and mutation analysis
Tissue microarrays were constructed from representative parts of the formalin-fixed paraffin-embedded tumour tissues using either a 0.7 mm diameter needle (the western Sweden series) or a 1 mm diameter needle (the SSGXVIII/AIO series). SLUG expression was evaluated using immunohistochemistry in 5 μm thick tissue microarray sections. Before staining, endogenous peroxidase activity was blocked by incubating deparaffinized tissue in hydrogen peroxidase, and antigen retrieval was performed using sodium citrate (10 mmol l−1, pH 6.0) in an autoclave (at 120 °C for 2 min). The anti-SLUG antibody (dilution 1 : 50; clone C19G7, Cell Signaling Technology, Danvers, MA, USA) was diluted in a Normal antibody Diluent (Immunologic, Duiven, The Netherlands), and incubated on slides at +4 °C overnight. Binding of the primary antibody was detected and visualized by using the N-Histofine Simple Stain MAX PO (R) kit (Nichirei Biosciences Inc, Tokyo, Japan) and 3,3′-diaminobenzidine (ImmPACT DAB, Vector Laboratories, Burlingame, CA, USA) following the manufacturer's recommendations. The slides were counterstained with Mayerś hematoxylin. Histologically normal kidney tubule cells served as a positive control and kidney glomerular cells as a negative control for the staining. The samples were considered positive for SLUG expression when nuclear and/or cytoplasmic staining was present in >20% of the tumour cells. The immunostainings were analysed blinded without knowledge of the clinical or histopathological data. KIT-positivity and mutation status of the GIST samples was verified by immunohistochemistry (rabbit polyclonal CD117-antibody, code A4502, Dako, Glostrup, Denmark) and using PCR and bidirectional sequencing as described elsewhere (Nilsson et al, 2005b; Andersson et al, 2006; Wardelmann et al, 2006).
Cell lines and transfection
Two human GIST cell lines were kindly provided by Dr Jonathan A Fletcher (Harvard Medical School, Boston, Massachusetts, USA). GIST882 harbors a homozygous missense mutation in KIT exon 13 encoding a Lys642Glu (K642E) mutant oncoprotein (Tuveson et al, 2001). GIST48 was obtained from a patient whose GIST progressed on imatinib therapy, and harbors a KIT exon 11 V560D missense mutation leading to p.Val560Asp and a secondary exon 17 (kinase activation loop) missense mutation D820A leading to p.Asp820Ala (Bauer et al, 2006). The authenticity of the cell lines was confirmed by sequencing. Cells were cultured in a humidified 5% CO2 atmosphere at 37 °C in the RPMI 1640 medium (GIBCO, CA, USA), supplemented with 20% fetal bovine serum with 2% penicillin/streptomycin (GIBCO).
GIST882 and GIST48 cells were transfected using the Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA). The cells were serum starved for 6 h before transfection in an Opti-MEM Reduced Serum Medium (GIBCO). The transfections were done according to the manufacturer's instructions adding 5 pM of siRNAs onto the cells. Transfections were done with an ON-TARGET plus Human KIT siRNA and an ON-TARGET plus Human SNAI2 (SLUG) siRNAs, which are pools of target-specific siRNAs (Thermo Scientific Dharmacon, Rockford, IL, USA). The ON-Target plus Non-Targeting Pool (Thermo Scientific Dharmacon) was used as a negative control. Imatinib mesylate was purchased from Cayman chemicals (Ann Arbor, MI, USA).
Cell proliferation and cell death assays
GIST cell proliferation was studied with the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Roche Diagnostics, Indianapolis, IN, USA). For the assay, GIST882 cells were plated at a density of 10 000 cells per well and GIST48 cells at a density of 15 000 cells per well in 96-well plates. The assays were performed thrice, using eight replicate wells at each time point. Cell proliferation was analysed after adding 10 μl of the MTT reagent into each well and incubation at 37 °C for 4 h, after which the solubilization solution was added and the cells were incubated at 37 °C overnight. The plates were analysed with a Multiscan EX Microplate photometer (Thermo Scientific, Rockford, IL, USA) at the wavelength of 540 nm.
GIST cell death after exposure to KIT siRNA or SLUG siRNA was analysed with a Click-iT TUNEL Alexa Fluor 488 Imaging Assay (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. GIST882 cells were plated at a density of 10 000 cells per well and GIST48 cells at a density of 15 000 cells per well in 96-well plates. After 24 h of culture at 37 °C the cells were transfected with siRNAs as described above. The TUNEL assay was performed 96 h after siRNA transfections. Cells were photographed (magnification × 200) using an immunofluorescence microscope (Leica CTR6000, Leica microsystems, Bannockburn, IL, USA). Cell death was measured in three separate experiments by counting the cells within 21 (5, 8 and 8) photographed fields of the microscope. The frequency of dying cells was expressed as the average number of dying cells per one microscope field.
Western blotting
Cultured GIST882 and GIST48 cells were rinsed in PBS (Lonza, Walkersville, MD, USA) and scraped into a RIPA lysis buffer (Pierce Biotechnology, Rockford, IL, USA) containing a Pierce Protease and Phosphatase Inhibitor Tablet (Thermo Fisher Scientific Inc., Rockford, IL, USA), followed by sonication. Protein concentration was determined with the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific Inc.). Protein 10 μg of protein was separated using a gel electrophoresis and blotted onto an Immobilon-P Transfer Membrane (Millipore, Billerica, MA, USA), and stained with a polyclonal rabbit anti-c-KIT antibody (dilution 1 : 10000; clone A4502, Agilent Technologies Dako, Glostrup, Denmark), a polyclonal rabbit anti-SLUG antibody (dilution 1 : 1000; clone C19G7, Cell Signaling Technology), a polyclonal rabbit anti-β-actin antibody (dilution 1 : 10 000; Bethyl Laboratories, Montgomery, TX, USA) or a Apoptosis Western Blot Cocktail (Abcam, Cambridge, UK). Primary antibodies were detected with specific horseradish-labelled secondary antibodies and using the SuperSignal West Pico and Femto Chemiluminescent Substrate (Thermo Fisher Scientific Inc.), following manufacturer's instructions. The basal level of SLUG expression was significantly lower in GIST882 cells than in GIST48 cells. Protein expression was detected after diluting Femto Substrate 1 : 5 with Pico Substrate.
Statistical analysis
The frequency tables were analysed with the χ2 test or Fisher's exact test. The groups resulting from the TUNEL analysis were compared using the Mann–Whitney U-test. Cumulative survival was estimated with the Kaplan–Meier method, and survival between groups was compared using a univariable Cox proportional hazards model. The interaction between tumour SLUG expression and imatinib treatment in the SSGXVIII/AIO series was calculated using a Gaussian process model for conditional event probabilities. Gaussian process model can model nonlinear effects of the covariates and interactions between covariates, and thus allow inspection of whether SLUG has an interaction effect with imatinib treatment or other covariates (Joensuu et al, 2012b). Overall survival was calculated from the date of surgery to the date of death, censoring patients who were alive on the last follow-up date. Recurrence-free survival (RFS) was calculated from the date of randomisation to the date of GIST recurrence or to death, whenever death preceded recurrence, censoring the patients alive on the date of last follow-up. The P values are two-sided. The statistical calculations were done using the IBM SPSS Statistics package v. 22.0, or with GPstuff 4.6 (Vanhatalo et al, 2013).
Results
Associations between SLUG and clinical, histopathological and molecular features of GIST
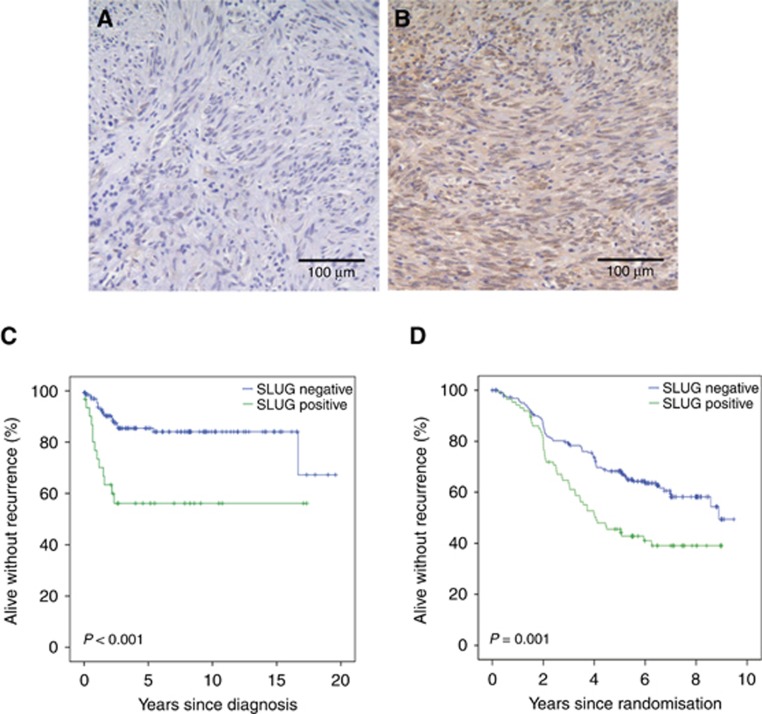
SLUG expression was present in 39 (20.9%) out of the 187 GISTs examined from the western Sweden series and in 86 (27.5%) out of the 313 GISTs in the SSGXVIII/AIO series (Figure 1A and B). Thus, SLUG was expressed in 125 (25.0%) out of the 500 GISTs that were immunostained for SLUG. The intracellular localisation of SLUG was analysed in the western Sweden series. SLUG was usually expressed both in the nucleus and the cytoplasm, and, therefore, any GIST SLUG expression, regardless of the subcellular location, was considered in further analyses.
Figure 1.
SLUG expression in GIST, and association with recurrence-free survival.An example of negative (A) and positive (B) expression of SLUG in GIST (magnification × 200). Association of tumour SLUG expression with recurrence-free survival in the western Sweden population-based series (C), and in the SSGXVIII/AIO series (D).
In the western Sweden series, where the patients did not receive adjuvant imatinib after surgery, SLUG expression was associated with a large tumour size at the time of the diagnosis, epithelioid type of tumour histology and nuclear pleomorphism, the presence of tumour necrosis, a high tumour mitotic count and presence of metastases at the time of GIST detection. No significant association was found with gender, age at the time of the diagnosis, tumour location in the gastrointestinal tract, or a high risk for recurrence when GISTs were classified according to the National Institute of Health Consensus stratification scheme (Fletcher et al, 2002) (Table 1). In the SSGXVIII/AIO series, where the patients were treated with surgery and adjuvant therapy and the patients had high-risk GIST, tumour SLUG expression was significantly associated with the presence of tumour rupture, whereas no association was found with the gender, age at the time of the diagnosis, tumour site, size or mitotic count, or the mutated gene (Table 2).
Table 1. Associations between GIST SLUG expression and 10 clinicopathological factors in the western Sweden series.
| Factor | SLUG negative n (%) | SLUG positive n (%) | P |
|---|---|---|---|
|
Age | |||
| Median (range) | 69 (35–92) | 69 (30–85) | 0.532 |
|
Gender | |||
| Female | 76 (81.7) | 17 (18.3) | 0.388 |
| Male | 72 (76.6) | 22 (23.4) | |
|
Tumour size (cm) | |||
| Median (range) | 5.0 (0.5–33.0) | 8.0 (1.0–35.0) | 0.002 |
|
Tumour site | |||
| Gastric | 82 (80.4) | 20 (19.6) | 0.645 |
| non-gastric | 66 (77.6) | 19 (22.4) | |
|
Mitotic count | |||
| <2 | 81 (89.0) | 10 (11.0) | 0.002 |
| 2–5 | 36 (72.0) | 14 (28.0) | |
| 6–10 | 15 (78.9) | 4 (21.1) | |
| >10 | 12 (54.5) | 10 (45.5) | |
| N.A. | 4 | 1 | |
|
Histology | |||
| Epithelioid | 11 (52.4) | 10 (47.6) | 0.001 |
| Spindle-like | 114 (86.4) | 18 (13.6) | |
| Mixed | 19 (73.1) | 7 (26.9) | |
| N.A. | 4 | 4 | |
|
Metastases at the time of diagnosis | |||
| Present | 12 (60.0) | 8 (40.0) | 0.039 |
| Not present | 136 (81.4) | 31 (18.6) | |
|
Nuclear pleomorphism | |||
| No or minimal | 40 (93.0) | 3 (7.0) | <0.0001 |
| Moderate | 87 (82.9) | 18 (17.1) | |
| Severe | 17 (53.1) | 15 (46.9) | |
| N.A. | 4 | 3 | |
|
Tumour necrosis | |||
| Not present | 76 (83.5) | 15 (16.5) | 0.003 |
| Present | 23 (59.0) | 16 (41.0) | |
| N.A. | 49 | 8 | |
|
Risk classification (NIH)a | |||
| Very low | 28 (90.3) | 3 (9.7) | 0.217 |
| Low | 46 (80.7) | 11 (19.3) | |
| Intermediate | 18 (90.0) | 2 (10.0) | |
| High | 44 (74.6) | 15 (25.4) | |
Abbreviations: N.A., not available; NIH, the National Institutes of Health.
Cancers with metastases at the time of the diagnosis (n=20) were excluded from the analysis.
Table 2. Associations between GIST SLUG expression and 7 clinicopathological factors in the SSGXVIII/AIO series.
| Factor | SLUG negative n (%) | SLUG positive n (%) | P |
|---|---|---|---|
|
Age | |||
| Median (range) | 61.5 (26–75) | 60 (22–83) | 0.806 |
|
Gender | |||
| Female | 113 (73.9) | 40 (26.1) | 0.606 |
| Male | 114 (71.3) | 46 (28.8) | |
|
Tumour size—cm | |||
| Median (range) | 9.7 (2.5–35.0) | 9.0 (1.5–30.0) | 0.930 |
|
Location | |||
| Gastric | 127 (75.1) | 42 (24.9) | 0.228 |
| Non-gastric | 98 (69.0) | 44 (31.0) | |
|
Mitotic count | |||
| <2 | 52 (82.5) | 11 (17.5) | 0.245 |
| 2–5 | 59 (71.1) | 24 (28.9) | |
| 6–10 | 32 (72.7) | 12 (27.3) | |
| >10 | 79 (68.7) | 36 (31.3) | |
| N.A. | 5 | 3 | |
|
Tumour rupture | |||
| No | 187 (75.1) | 62 (24.9) | 0.044 |
| Yes | 40 (62.5) | 24 (37.5) | |
|
Mutation site | |||
| KIT | 176 (72.4) | 67 (27.6) | |
| Exon 9 | 17 | 5 | |
| Exon11 | 156 | 60 | |
| Exon 13 | 3 | 2 | |
| PDGFRA | 25 (65.8) | 13 (34.2) | |
| Wild type for KIT and PDGFRA | 19 (79.2) | 5 (20.8) | 0.506 |
Abbreviations: HPF, high power field of a microscope; N.A., not available; PDGFRA, platelet-derived growth factor receptor alpha gene.
Associations with survival
Patients whose GIST expressed SLUG had less favorable RFS than patients whose GIST did not express SLUG in the western Sweden series (HR=3.40, 95% CI=1.67–6.89, P=0.001; Figure 1C). Similarly, overall survival of these patients was less favorable (HR=1.88, 95% CI=1.20–2.93, P=0.006). When SLUG expression (positive vs negative) was entered as a covariable into a Cox multivariable analysis together with tumour mitotic count (⩽5 vs >5 mitosis/50 HPFs), tumour site (non-gastric vs gastric), and tumour size (as a continuous variable) using RFS as the endpoint, only tumour size (HR=1.09, 95% CI=1.03–1.15, P=0.002) and mitotic count (HR=10.64, 95% CI=4.13–27.38, P<0.001) were significantly associated with survival, whereas non-gastric tumour site (HR=1.90, 95% CI=0.91–3.96, P>0.089) and SLUG expression (HR=1.55, 95% CI=0.66–3.67, P>0.319) were not. Similarly, SLUG expression in the high risk group associated with poor RFS (HR=1.76, 95% CI=1.06–2.92, P=0.029) but was not independent prognostic factor in Cox multivariable analysis (data not shown). Tumour rupture data were not available; the 20 patients who had distant metastases at the time of the diagnosis and four patients whose mitotic count was not available were excluded from the analysis.
In accordance with these observations, in the SSGXVIII/AIO series patients with SLUG-positive tumour had inferior RFS as compared to those whose GIST did not express SLUG (HR=1.83, 95% CI 1.29–2.60; P=0.001; Figure 1D; Supplementary Figure 1). There was no significant interaction between tumour SLUG expression and the duration of adjuvant imatinib treatment. GIST SLUG expression was associated with poor RFS also in the subset of patients with KIT exon 11 deletion or insertion-deletion mutation (n=136, the largest single genotypic subgroup, of which 42 930.9%) stained positively for SLUG; HR=2.22, 95% CI 1.30–3.80; P=0.004), whereas SLUG expression was not significantly associated with RFS among patients with KIT exon 11 substitution mutation (n=59, the second largest genotypic subgroup, of which 14 [23.7%] had positive staining for SLUG; HR=1.56, 95% CI 0.58–4.24; P=0.379). Tumour SLUG expression was independently associated with GIST patient RFS when it was entered into a Cox multivariable analysis as a covariable together with the four established prognostic factors in GIST (tumour mitotic count, tumour site, tumour rupture and tumour size) and the duration of adjuvant imatinib (1 year or 3 years) in the SSGXVIII/AIO series (HR=1.63, 96% CI 1.13.2.36, P=0.009; Table 3). However, each of the four standard factors and the duration of adjuvant imatinib predicted GIST recurrence more strongly than tumour SLUG expression (P=0.001 or smaller for each covariable, Table 3). The results of the Cox multivariable model were essentially similar and tumour SLUG expression remained an independent factor when tumour mitotic count was entered into the model categorised as shown in Table 2 in place of a continuous covariable.
Table 3. Influence of six factors on recurrence-free survival in a Cox multivariable model in the SSGXVIII/AIO series.
| Variable | Hazard ratio (95% CI) | P |
|---|---|---|
| Tumour mitotic count | 1.03 (1.02–1.03) | <0.001 |
| As a continuous variable | ||
| Tumour site | 3.03 (2.05–4.46) | <0.001 |
| Non-gastric vs gastric | ||
| Duration of adjuvant imatinib | 2.22 (1.52–3.22) | <0.001 |
| 1 year vs 3 years | ||
| Tumour rupture | 2.25 (1.52–3.33) | <0.001 |
| Present vs absent | ||
| Tumour size | 1.05 (1.02–1.07) | 0.001 |
| As a continuous variable | ||
| Tumour SLUG expression | 1.63 (1.13–2.36) | 0.009 |
| Present vs absent |
Abbreviation: CI, confidence interval.
SLUG and KIT silencing in GIST cell lines
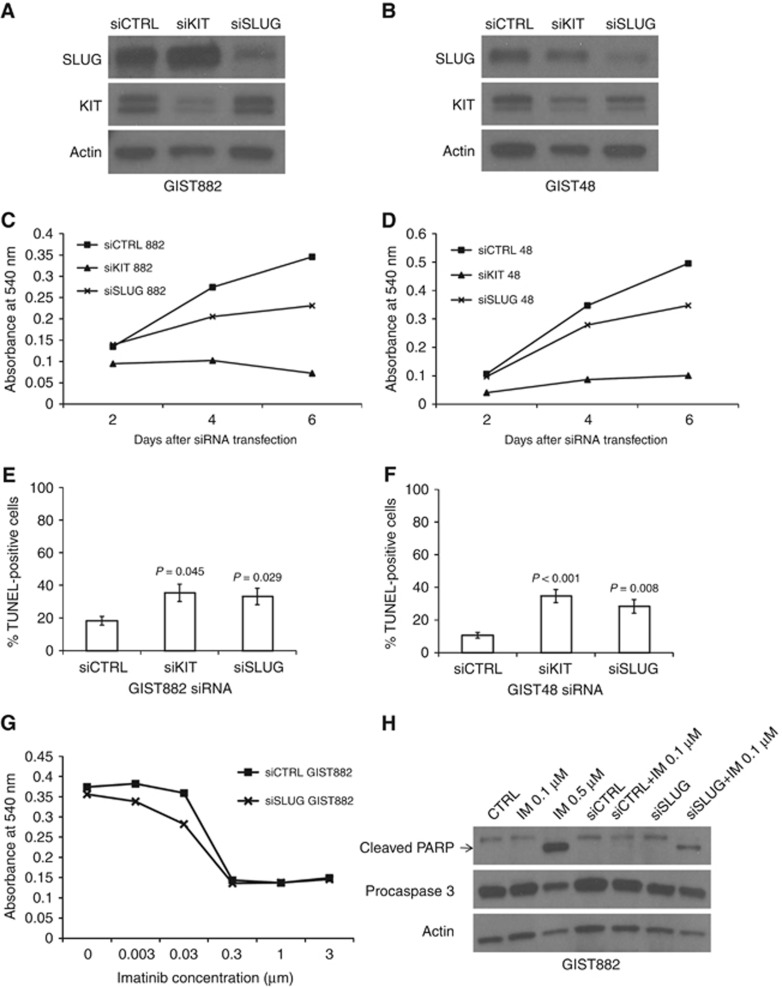
The effect of SLUG silencing was investigated in the GIST48 and GIST882 cell lines. Both cell lines expressed SLUG, but expression was substantially weaker in GIST882 as compared with GIST48 (Figure 2A and B). As tumour SLUG expression was associated with a high GIST cell proliferation rate in the western Sweden series, we studied whether SLUG promotes cell viability in GIST48 and GIST882 cell lines using the MTT assay. Blocking of SLUG expression by RNA interference reduced cell proliferation in both cell lines during six days of culture (P<0.001 for each cell line, standard error (s.e.) in all measured time points<±0.01; Figure 2C and D). Downregulation of KIT or SLUG with siRNAs increased cell death in both cell lines when the numbers of dying cells were measured using the TUNEL assay 96 h after treatment with the siRNAs (Figure 2E and F). In the GIST882 cell line SLUG siRNA transfection increased the mean proportion of DNA fragmentation in nuclei from 18.3% (s.e.=±2.6%) in the control siRNA to 33.2% (s.e.=±5.1%, P=0.029) and KIT siRNA to 35.4% (s.e.=±5.3%, P=0.045). Similarly, in the GIST48 cell line the mean proportion of dying cells increased from 10.7% (s.e.=1.8%) in the control to 28.4% (s.e.=±4.2%, P=0.008) and 34.7% (s.e.=±4.0%, P <0.001) after treating the cells with SLUG siRNA and KIT siRNA, respectively. In addition, downregulation of SLUG showed synergy with imatinib on GIST882 cell line proliferation and sensitised cells to imatinib-induced apoptosis as measured with presence of caspase cleaved PARP (Figure 2G and H), although synergistic effect was not found in GIST48 cell line (data not shown).
Figure 2.
SLUG and KIT siRNA silencing in GIST cell lines.An example of SLUG, KIT and actin expression after siRNA transfection in the GIST882 (A) and the GIST48 (B) cell line. Cell proliferation decreased (C, D) and the rate of cell death (E, F) increased in the GIST882 cell line and in the GIST48 cell line after transfection with KIT or SLUG siRNA. An efficacy of imatinib in the GIST882 cell line was enhanced by the downregulation of SLUG expression (G). The simultaneous downregulation of SLUG expression and KIT inhibition sensitised GIST882 cells to lower imatinib concentration (H).
Discussion
We found that tumour SLUG expression was present in a quarter of GISTs and that its expression was associated with unfavourable RFS both in a patient population treated with surgery alone and in those treated with surgery and adjuvant imatinib. In line with the adverse influence of SLUG expression on survival outcomes, SLUG expression was associated with a few known risk factors for GIST recurrence and metastasis, such as a high cell proliferation rate, presence of tumour rupture, and presence of distant metastases at the time of the diagnosis. However, tumour SLUG expression was not associated with non-gastric location of the primary tumour, which is considered an established adverse prognostic factor in GIST (Joensuu et al, 2016). Blocking of SLUG expression using siRNA reduced cell proliferation and increased cell death in vivo in the two GIST cell lines investigated.
To our knowledge, this is the first report concerning the clinical significance of SLUG expression in GIST. Prior studies, performed in patient populations such as oesophageal cancer, colorectal cancer and lung cancer, found that patients with SLUG-positive cancer have poor outcomes also in these types of human cancer (Uchikado et al, 2005; Shioiri et al, 2006; Wu et al, 2015). Taken together, these observations suggest that tumour SLUG expression may be an adverse prognostic feature in several types of human cancer, including some sarcomas. In the present series, tumour SLUG expression was a relatively strong adverse factor in univariable survival analyses and had independent prognostic value in a multivariable analysis of the SSGXVIII/AIO series. On the other hand, a majority of even high-risk GISTs did not express SLUG suggesting that elevated SLUG expression is not related to cancer progression.
To understand better the adverse effect of SLUG on GIST patient outcome, we studied SLUG in the two GIST cell lines. These cell lines had distinct basal SLUG expression levels, and SLUG downregulation decreased cell viability and increased cell death in both cell lines in accordance with studies performed in other experimental tumour models (Wu et al, 2005; Vitali et al, 2008; Emadi Baygi et al, 2010). SLUG may antagonise apoptosis by regulation of caspases or repressing expression of the pro-apoptotic PUMA protein (Tribulo et al, 2004; Wu et al, 2005). However, we did not find any effect on PUMA expression in GIST cell lines when SLUG expression was repressed using siRNA (data not shown). SLUG silencing sensitises cholangiocarcinoma and ovarian cancer cells to cisplatin (Zhang et al, 2011a; Haslehurst et al, 2012), and SLUG may have an essential role in a signalling network that drives survival and imatinib resistance of Bcr-Abl-expressing cells (Mancini et al, 2010). As a transcription factor, SLUG may mediate cell survival and drug resistance in cancer cells by regulation set of genes functioning in various signalling pathways. Theoretically, imatinib mesylate and SLUG down-regulation could be combined to treat some malignancies such as GIST (Vitali et al, 2008; Mancini et al, 2010), but the means to deliver effective anti-SLUG therapy efficiently and safely are currently lacking. It remains to be seen whether emerging technologies such as the RNA silencing technology or target proteins to proteolysis targeting chimeras (PROTACs) will resolve this problem (Deshaies, 2015).
SLUG expression was associated with unfavourable survival outcomes in both series in univariable analyses, but it emerged as an independent factor in the SSGXVIII/AIO trial series only. This might suggest that SLUG is a predictive factor rather than a prognostic factor in GIST, but such a conclusion might be premature and requires confirmation from other series. The patient populations differ between the series as the western Sweden GIST series is a population-based series that includes both low-risk and high-risk patients, whereas the SSGXVIII series included patients with a high estimated risk for recurrence. The covariables in the Cox models may also differ.
The present study has some limitations. The clinical series investigated are large and the treatment given could be taken into account in the analyses, but the quantitation of SLUG expression from immunostained slides is somewhat subjective, and we could not control factors that may influence SLUG detection by immunohistochemistry, such as the methods and the times of tissue fixation. SLUG expression or nuclear localisation in some tumours might be missed from analysis of tissue microarray samples, as SLUG expression may vary within tumour tissue. However, we found significant differences in patient outcome when only the nuclear expression or both the cytoplasmic and the nuclear expression were considered in survival analyses (data not shown). Finally, there are only few GIST cell lines available, and we had access to only two of these despite our efforts to obtain more cell lines for the study. Although the findings from the two cell lines are supportive to the data from the clinical series, they should be viewed with some caution, as the effects of SLUG might vary between GISTs and the cell lines derived from them.
In sum, the results suggest that about a quarter of GISTs express SLUG and that its expression is associated with unfavourable RFS of GIST patients. The results suggest that SLUG may function as a pro-survival factor in GIST.
Acknowledgments
This study was supported by Academy of Finland, Cancer Society of Finland, Jane and Erkko Foundation, HUCH Research Funds, Sigrid Juselius Foundation, Luise and Henrik Kuningas Foundation, and Ida Montin Foundation.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
The authors declare no conflict of interest.
Supplementary Material
References
- Andersson J, Bumming P, Meis-Kindblom JM, Sihto H, Nupponen N, Joensuu H, Oden A, Gustavsson B, Kindblom LG, Nilsson B (2006) Gastrointestinal stromal tumours with KIT exon 11 deletions are associated with poor prognosis. Gastroenterology 130: 1573–1581. [DOI] [PubMed] [Google Scholar]
- Bauer S, Yu LK, Demetri GD, Fletcher JA (2006) Heat shock protein 90 inhibition in imatinib-resistant gastrointestinal stromal tumour. Cancer Res 66: 9153–9161. [DOI] [PubMed] [Google Scholar]
- Bermejo-Rodriguez C, Perez-Caro M, Perez-Mancera PA, Sanchez-Beato M, Piris MA, Sanchez-Garcia I (2006) Mouse cDNA microarray analysis uncovers Slug targets in mouse embryonic fibroblasts. Genomics 87: 113–118. [DOI] [PubMed] [Google Scholar]
- Catalano A, Rodilossi S, Rippo MR, Caprari P, Procopio A (2004) Induction of stem cell factor/c-Kit/slug signal transduction in multidrug-resistant malignant mesothelioma cells. J Biol Chem 279: 46706–46714. [DOI] [PubMed] [Google Scholar]
- Cobaleda C, Perez-Caro M, Vicente-Duenas C, Sanchez-Garcia I (2007) Function of the zinc-finger transcription factor SNAI2 in cancer and development. Annu Rev Genet 41: 41–61. [DOI] [PubMed] [Google Scholar]
- Dematteo RP, Ballman KV, Antonescu CR, Maki RG, Pisters PW, Demetri GD, Blackstein ME, Blanke CD, von Mehren M, Brennan MF, Patel S, McCarter MD, Polikoff JA, Tan BR, Owzar K American College of Surgeons Oncology Group (ACOSOG) Intergroup Adjuvant GIST Study Team (2009) Adjuvant imatinib mesylate after resection of localised, primary gastrointestinal stromal tumour: a randomised, double-blind, placebo-controlled trial. Lancet 373: 1097–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetri GD, von Mehren M, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, Heinrich MC, Tuveson DA, Singer S, Janicek M, Fletcher JA, Silverman SG, Silberman SL, Capdeville R, Kiese B, Peng B, Dimitrijevic S, Druker BJ, Corless C, Fletcher CD, Joensuu H (2002) Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumours. N Engl J Med 347: 472–480. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ (2015) Protein degratation: prime time for PROTACs. Nat Chem Biol 11: 634–635. [DOI] [PubMed] [Google Scholar]
- Ducimetiere F, Lurkin A, Ranchere-Vince D, Decouvelaere AV, Peoc'h M, Istier L, Chalabreysse P, Muller C, Alberti L, Bringuier PP, Scoazec JY, Schott AM, Bergeron C, Cellier D, Blay JY, Ray-Coquard I (2011) Incidence of sarcoma histotypes and molecular subtypes in a prospective epidemiological study with central pathology review and molecular testing. PLoS One 6: e20294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emadi Baygi M, Soheili ZS, Essmann F, Deezagi A, Engers R, Goering W, Schulz WA (2010) Slug/SNAI2 regulates cell proliferation and invasiveness of metastatic prostate cancer cell lines. Tumour Biol 31: 297–307. [DOI] [PubMed] [Google Scholar]
- Fletcher CD, Berman JJ, Corless C, Gorstein F, Lasota J, Longley BJ, Miettinen M, O'Leary TJ, Remotti H, Rubin BP, Shmookler B, Sobin LH, Weiss SW (2002) Diagnosis of gastrointestinal stromal tumours: a consensus approach. Hum Pathol 33: 459–465. [DOI] [PubMed] [Google Scholar]
- Haslehurst AM, Koti M, Dharsee M, Nuin P, Evans K, Geraci J, Childs T, Chen J, Li J, Weberpals J, Davey S, Squire J, Park PC, Feilotter H (2012) EMT transcription factors snail and slug directly contribute to cisplatin resistance in ovarian cancer. BMC Cancer 12: 91–2407-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Blanke CD, Demetri GD, Joensuu H, Roberts PJ, Eisenberg BL, von Mehren M, Fletcher CD, Sandau K, McDougall K, Ou WB, Chen CJ, Fletcher JA (2006) Molecular correlates of imatinib resistance in gastrointestinal stromal tumours. J Clin Oncol 24: 4764–4774. [DOI] [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A, Demetri GD, Fletcher CD, Fletcher JA (2003) PDGFRA activating mutations in gastrointestinal stromal tumours. Science 299: 708–710. [DOI] [PubMed] [Google Scholar]
- Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, Muhammad Tunio G, Matsuzawa Y, sKanakura Y, Shinomura Y, Kitamura Y (1998) Gain-of-function mutations of c-kit in human gastrointestinal stromal tumours. Science 279: 577–580. [DOI] [PubMed] [Google Scholar]
- Joensuu H (2008) Risk stratification of patients diagnosed with gastrointestinal stromal tumour. Hum Pathol 39: 1411–1419. [DOI] [PubMed] [Google Scholar]
- Joensuu H, Eriksson M, Sundby Hall K, Hartmann JT, Pink D, Schutte J, Ramadori G, Hohenberger P, Duyster J, Al-Batran SE, Schlemmer M, Bauer S, Wardelmann E, Sarlomo-Rikala M, Nilsson B, Sihto H, Monge OR, Bono P, Kallio R, Vehtari A, Leinonen M, Alvegard T, Reichardt P (2012. a) One vs three years of adjuvant imatinib for operable gastrointestinal stromal tumour: a randomized trial. J Am Med Assoc 307: 1265–1272. [DOI] [PubMed] [Google Scholar]
- Joensuu H, Eriksson M, Sundby Hall K, Reichardt A, Hartmann JT, Pink D, Ramadori G, Hohenberger P, Al-Batran SE, Schlemmer M, Bauer S, Wardelmann E, Nilsson B, Sihto H, Bono P, Kallio R, Junnila J, Alvegard T, Reichardt P (2016) Adjuvant Imatinib for high-risk GI stromal tumour: analysis of a randomized trial. J Clin Oncol 34: 244–250. [DOI] [PubMed] [Google Scholar]
- Joensuu H, Vehtari A, Riihimaki J, Nishida T, Steigen SE, Brabec P, Plank L, Nilsson B, Cirilli C, Braconi C, Bordoni A, Magnusson MK, Linke Z, Sufliarsky J, Federico M, Jonasson JG, Dei Tos AP, Rutkowski P (2012. b) Risk of recurrence of gastrointestinal stromal tumour after surgery: an analysis of pooled population-based cohorts. Lancet Oncol 13: 265–274. [DOI] [PubMed] [Google Scholar]
- Kim S, Yao J, Suyama K, Qian X, Qian BZ, Bandyopadhyay S, Loudig O, De Leon-Rodriguez C, Zhou ZN, Segall J, Macian F, Norton L, Hazan RB (2014) Slug promotes survival during metastasis through suppression of Puma-mediated apoptosis. Cancer Res 74: 3695–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurrey NK, Jalgaonkar SP, Joglekar AV, Ghanate AD, Chaskar PD, Doiphode RY, Bapat SA (2009) Snail and slug mediate radioresistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem Cells 27: 2059–2068. [DOI] [PubMed] [Google Scholar]
- Mancini M, Petta S, Iacobucci I, Salvestrini V, Barbieri E, Santucci MA (2010) Zinc-finger transcription factor slug contributes to the survival advantage of chronic myeloid leukemia cells. Cell Signal 22: 1247–1253. [DOI] [PubMed] [Google Scholar]
- Motro B, van der Kooy D, Rossant J, Reith A, Bernstein A (1991) Contiguous patterns of c-kit and steel expression: analysis of mutations at the W and Sl loci. Development 113: 1207–1221. [DOI] [PubMed] [Google Scholar]
- Nannini M, Astolfi A, Urbini M, Indio V, Santini D, Heinrich MC, Corless CL, Ceccarelli C, Saponara M, Mandrioli A, Lolli C, Ercolani G, Brandi G, Biasco G, Pantaleo MA (2014) Integrated genomic study of quadruple-WT GIST (KIT/PDGFRA/SDH/RAS pathway wild-type GIST). BMC Cancer 14: 685–2407-14-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA (2002) The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol 3: 155–166. [DOI] [PubMed] [Google Scholar]
- Nilsson B, Bumming P, Meis-Kindblom JM, Oden A, Dortok A, Gustavsson B, Sablinska K, Kindblom LG (2005. a) Gastrointestinal stromal tumours: the incidence, prevalence, clinical course, and prognostication in the preimatinib mesylate era–a population-based study in western Sweden. Cancer 103: 821–829. [DOI] [PubMed] [Google Scholar]
- Nilsson B, Bumming P, Meis-Kindblom JM, Oden A, Dortok A, Gustavsson B, Sablinska K, Kindblom LG (2005. b) Gastrointestinal stromal tumours: the incidence, prevalence, clinical course, and prognostication in the preimatinib mesylate era–a population-based study in western Sweden. Cancer 103: 821–829. [DOI] [PubMed] [Google Scholar]
- Perez-Losada J, Sanchez-Martin M, Perez-Caro M, Perez-Mancera PA, Sanchez-Garcia I (2003) The radioresistance biological function of the SCF/kit signaling pathway is mediated by the zinc-finger transcription factor Slug. Oncogene 22: 4205–4211. [DOI] [PubMed] [Google Scholar]
- Perez-Losada J, Sanchez-Martin M, Rodriguez-Garcia A, Sanchez ML, Orfao A, Flores T, Sanchez-Garcia I (2002) Zinc-finger transcription factor Slug contributes to the function of the stem cell factor c-kit signaling pathway. Blood 100: 1274–1286. [PubMed] [Google Scholar]
- Qian J, Liu H, Chen W, Wen K, Lu W, Huang C, Fu Z (2013) Knockdown of Slug by RNAi inhibits the proliferation and invasion of HCT116 colorectal cancer cells. Mol Med Rep 8: 1055–1059. [DOI] [PubMed] [Google Scholar]
- Sanchez-Martin M, Perez-Losada J, Rodriguez-Garcia A, Gonzalez-Sanchez B, Korf BR, Kuster W, Moss C, Spritz RA, Sanchez-Garcia I (2003) Deletion of the SLUG (SNAI2) gene results in human piebaldism. Am J Med Genet A 122A: 125–132. [DOI] [PubMed] [Google Scholar]
- Shioiri M, Shida T, Koda K, Oda K, Seike K, Nishimura M, Takano S, Miyazaki M (2006) Slug expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Br J Cancer 94: 1816–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spritz RA, Beighton P (1998) Piebaldism with deafness: molecular evidence for an expanded syndrome. Am J Med Genet 75: 101–103. [DOI] [PubMed] [Google Scholar]
- Tribulo C, Aybar MJ, Sanchez SS, Mayor R (2004) A balance between the anti-apoptotic activity of Slug and the apoptotic activity of msx1 is required for the proper development of the neural crest. Dev Biol 275: 325–342. [DOI] [PubMed] [Google Scholar]
- Tuveson DA, Willis NA, Jacks T, Griffin JD, Singer S, Fletcher CD, Fletcher JA, Demetri GD (2001) STI571 inactivation of the gastrointestinal stromal tumour c-KIT oncoprotein: biological and clinical implications. Oncogene 20: 5054–5058. [DOI] [PubMed] [Google Scholar]
- Uchikado Y, Natsugoe S, Okumura H, Setoyama T, Matsumoto M, Ishigami S, Aikou T (2005) Slug Expression in the E-cadherin preserved tumours is related to prognosis in patients with esophageal squamous cell carcinoma. Clin Cancer Res 11: 1174–1180. [PubMed] [Google Scholar]
- Vanhatalo J, Riihimäki J, Hartikainen J, Jylänki P, Tolvanen V, Vehtari A (2013) GPstuff: Bayesian Modeling with Gaussian Processes. JMLR 1175–1179.
- Vitali R, Mancini C, Cesi V, Tanno B, Mancuso M, Bossi G, Zhang Y, Martinez RV, Calabretta B, Dominici C, Raschella G (2008) Slug (SNAI2) down-regulation by RNA interference facilitates apoptosis and inhibits invasive growth in neuroblastoma preclinical models. Clin Cancer Res 14: 4622–4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shi J, Chai K, Ying X, Zhou BP (2013) The Role of Snail in EMT and Tumourigenesis. Curr Cancer Drug Targets 13: 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardelmann E, Merkelbach-Bruse S, Pauls K, Thomas N, Schildhaus HU, Heinicke T, Speidel N, Pietsch T, Buettner R, Pink D, Reichardt P, Hohenberger P (2006) Polyclonal evolution of multiple secondary KIT mutations in gastrointestinal stromal tumours under treatment with imatinib mesylate. Clin Cancer Res 12: 1743–1749. [DOI] [PubMed] [Google Scholar]
- Wu DW, Lee MC, Hsu NY, Wu TC, Wu JY, Wang YC, Cheng YW, Chen CY, Lee H (2015) FHIT loss confers cisplatin resistance in lung cancer via the AKT/NF-kappaB/Slug-mediated PUMA reduction. Oncogene 34: 2505–2515. [DOI] [PubMed] [Google Scholar]
- Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, Look AT (2005) Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell 123: 641–653. [DOI] [PubMed] [Google Scholar]
- Yang HW, Menon LG, Black PM, Carroll RS, Johnson MD (2010) SNAI2/Slug promotes growth and invasion in human gliomas. BMC Cancer 10: 301-2407-10-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Chen D, Wang X, Zhang S, Wang J, Gao Y, Yan B (2011. a) RNA interference targeting slug increases cholangiocarcinoma cell sensitivity to cisplatin via upregulating PUMA. Int J Mol Sci 12: 385–400. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang K, Zhang S, Jiao X, Wang H, Zhang D, Niu Z, Shen Y, Lv L, Zhou Y (2011. b) Slug regulates proliferation and invasiveness of esophageal adenocarcinoma cells in vitro and in vivo. Med Oncol 28: 1089–1100. [DOI] [PubMed] [Google Scholar]
- Zhou W, Lv R, Qi W, Wu D, Xu Y, Liu W, Mou Y, Wang L (2014) Snail contributes to the maintenance of stem cell-like phenotype cells in human pancreatic cancer. PLoS One 9: e87409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.