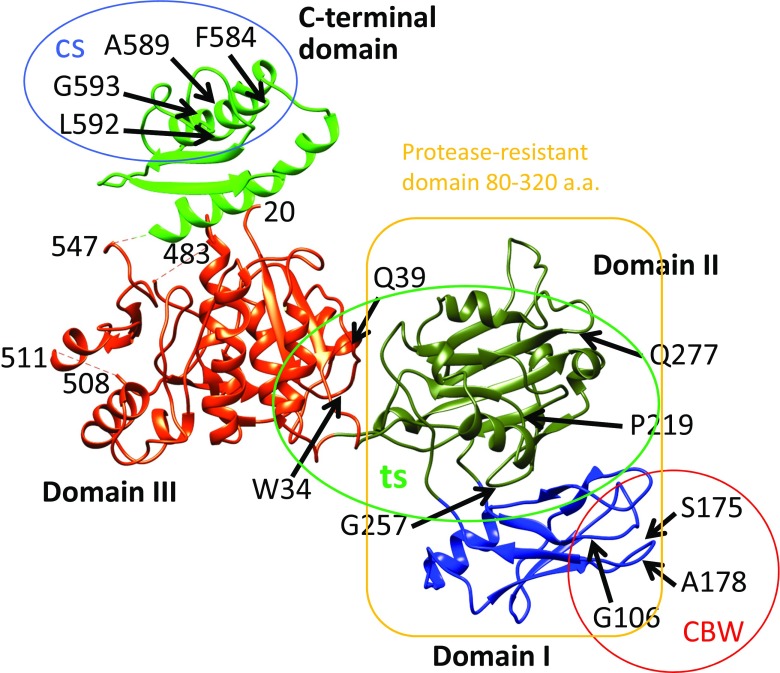
Fig. 6.
Crystal structure of gp18 (residues 20–510) (Aksyuk et al. 2009) in combination with the predicted C-terminal domain (Fokine et al. 2013). Four domains of gp18 are shown in blue (Domain I), olive green (Domain II), orange–red (Domain III) and green (C-terminal domain). The N-terminal 20 residues and residues 484–496 are not ordered in the crystal structure and thus not observed. Residue 510 is the last residue in the crystal structure. Mutation sites with a number of phenotypes were mapped by DNA sequencing of the mutants (Takeda et al. 2004). The temperature-sensitive mutants (ts; grow at 30 °C, but not at 42 °C) were mapped as W34R (or P), Q39P, P219L, G257D, Q277Y (or C, R, F), W487R, G587N and Q589Y. The heat-sensitive mutants (hs; lose viability upon incubation for 30 min at 55 °C) were mapped as P5S, R565C and G598S. The cold-sensitive mutants (cs; grow at 37 °C, but not at 25 °C) were mapped as R584H, Q589E and Q592E (or R); they are in the C-terminal domain circled). The carbowax mutants (CBW; can infect in the presence of a high concentration of polyethylene glycol) were mapped at G106S, S175F and A178V, in Domain I, and the mutation sites are circled