Fig. 1.
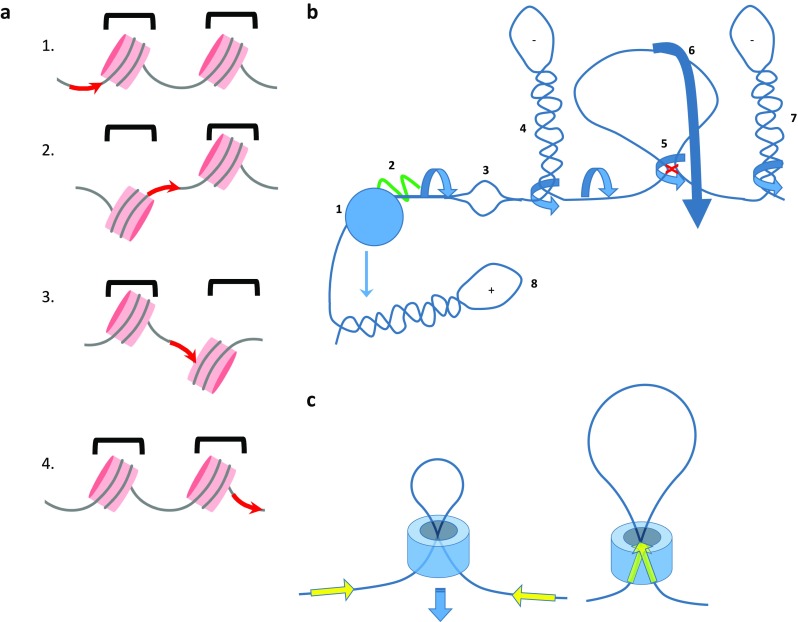
Static, constrained, and dynamic supercoils are context-dependent. a Pink disks indicate nucleosomes. Each nucleosome includes approximately ∼1.7 left-handed (negative) wraps of DNA that due to slight over-twisting (positive) of the helical axis yields a net −1.05 constrained supercoils/nucleosome. 1, 2 A dynamic pulse of torque pumped into the DNA (red arrow) must be accommodated either by propagation through the DNA, disrupting DNA–histone interactions (1), or by an en bloc rotation of the nucleosome that transmits the stress in a salutatory manner to the subsequent linker (2). 3, 4 In a similar manner the dynamic torsional stress is propagated along the chromatin fiber, etc. b The dynamic transmission of torsional stress is likely to be highly context-sensitive. Counter-rotation of RNA polymerase II (RNAPII) (1) and the template pumps torque into the chromatin fiber and—unless the RNAP is immobilized—may entwine the nascent RNA (2, green). Upstream negative supercoils may focally melt DNA (3) that can then adopt other non-B DNA conformations. Torsional stress may extrude plectonemes (4, 7) that compete for formation and growth in a static situation (the dynamics in vivo have not been described). A loop between DNA-bound upstream factors (red x) creates a topological domain insulated from dynamic supercoils (5); in such a situation a rotation of the entire loop about a stem (5) would create a plectoneme from segments embracing the domain, or a translational rotation (6) would transmit the stress as twist to the adjacent segment. If the loop were too encumbered to rotate or translocate, it would comprise a true topological boundary confining the torsional stress. c A chromatin loop can be contained within a collar that creates a topographic, but not true topological domain. For example, sliding of a cohesin (blue) ring along chromatin could help to juxtapose CTCF sites (yellow arrow) at the boundaries of a topologically associated domain. Whether the molecular architecture at these CTCF sites fixes linking number to define a topological domain has not yet been established
