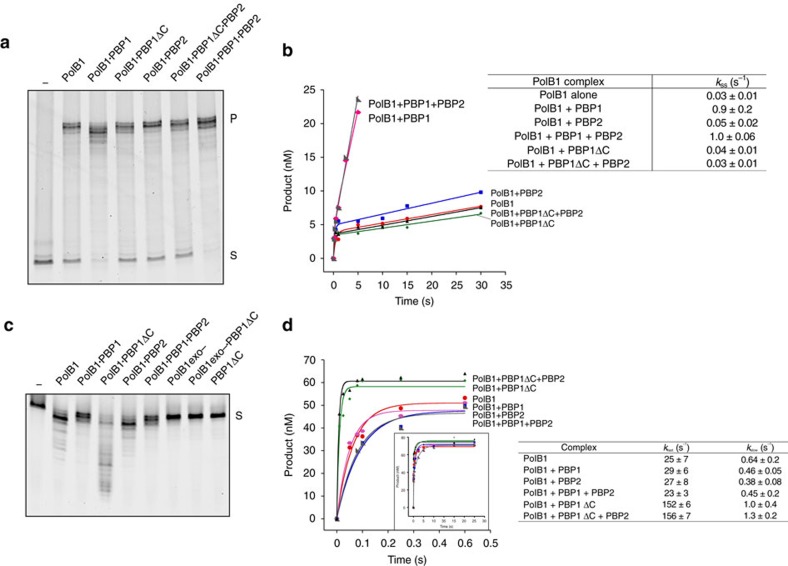
Figure 6. Activity assays for PolB1 assemblies.
(a) Primer-extension activities of PolB1-HE and sub-assemblies. Reactions contained 2.5 nM enzyme and 50 nM DNA substrate. Reactions were incubated at 75 °C for 5 min. Reaction products were analysed by denaturing PAGE. ‘S' denotes substrate and ‘P' denotes full-length product. (b) Steady-state dGTP Incorporation Kinetics for PolB1-HE and sub-assemblies. 5′-FAM-labelled 50 nt primer annealed to template DNA was pre-incubated with polymerase and reacted with dGTP using rapid quench flow (RQF) instrumentation from 0.1–30 s and quenched with 100 mM EDTA. 51 nt dGTP incorporation product was resolved from 50 nt substrate using capillary electrophoresis. 51 nt product was graphed as a function of time to the steady-state burst equation ([Product]=A[1-exp(−kobst)+k2t]) to obtain a kss (k2/A) for PolB1 (circle), PolB1 + PBP1 (hexagon), PolB1+PBP2 (square), PolB1-HE (right-angled triangle), PolB1+PBP1ΔC (diamond),PolB1+PBP1ΔC+PBP2 (triangle) (left panel). The kinetic parameters for PolB1 sub-assemblies are indicated on the right panel. (c) Exonuclease activities of PolB1-HE and sub-assemblies. Reactions contained 1 μM PolB1-HE and subcomplexes with 1 μM DNA substrate. Reaction products were analysed by denaturing PAGE. ‘PBP1ΔC' denotes PBP1 C-terminal tail deletion mutant, ‘PolB1exo-' denotes PolB1 exo-nuclease deficient mutant. (d) Pre-Steady-State Exonuclease Kinetics for PolB1 Complexes. 5′-FAM-labelled 50 nt exonuclease oligonucleotide containing a 3′ dGMP was incubated with polymerase using RQF instrumentation from 0.01–30 s and quenched with 1 N H2SO4+0.25% SDS. 49 nt exonuclease product was resolved from 50 nt substrate using capillary electrophoresis. 49 nt product was graphed as a function of time to a double-exponential equation ([Product]=A1[1-exp(−kfastt)]+A2[1-exp(−kslowt)]) to obtain a kfast and kslow for dGMP removal by PolB1 (circle), PolB1+PBP1 (hexagon), PolB1+PBP2 (square), PolB1-HE (right-angled triangle), PolB1+PBP1ΔC (diamond), PolB1+PBP1ΔC+PBP2 (triangle). The kinetic parameters for PolB1 sub-assemblies are indicated on the right panel.