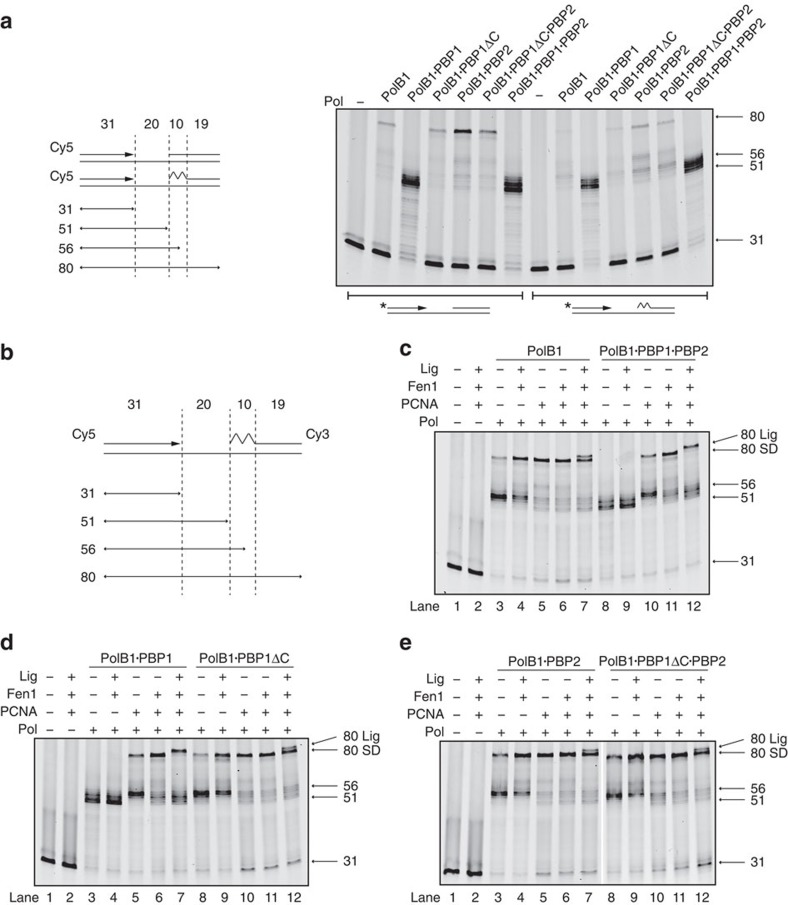
Figure 7. Strand displacement and lagging strand maturation activity of PolB1-HE and sub-assemblies.
(a) Strand-displacement activity of PolB1-containing holoenzyme and sub-assemblies. Substrate information is illustrated on the left. Primer was labelled with Cy5, jagged region represents the ribonucleotide primer. Reactions contained 10 nM PolB1 sub-assemblies and 50 nM DNA. Reaction products were analysed by denaturing PAGE. Notable product sizes are indicated on the right of the gel. (b) Diagram of the substrate used in the following experiments. The primer was labelled with Cy5 and downstream RNA-DNA hybrid oligonucleotide was labelled with Cy3. Lengths of all the oligos and possible reaction products are labelled. The jagged region represents the RNA component of the downstream ‘Okazaki fragment'. (c–e) Lagging strand maturation assay for all six PolB1 sub-assemblies. Reactions contained 12.5 nM PolB1 sub-assemblies, 200 nM PCNA, 50 nM Fen1, 400 nM Lig1, and 50 nM double-labelled substrate. Reaction products were analysed by denaturing PAGE. The gels were scanned twice with the Cy5 channel (shown in the figure) showing polymerase/strand-displacement products and Cy3 channel showing Fen1-mediated degradation and Lig1 catalyzed ligation products (presented as Supplementary Fig. 8c–e). Product sizes are indicated on the right of the gel. Full strand-displacement product (80 SD) and the ligation product (80 Lig) are also indicated on the right of the gel.