Table 1. Data collection and refinement statistics for PBP2 (SSO6202).
| Native | Gadolinium derivative | |||
|---|---|---|---|---|
| Data collection | ||||
| Space group | P3221 | P3221 | ||
| Cell dimensions | ||||
| a, b, c (Å) | 61.8, 61.8, 35.4 | 62.1, 62.1, 35.7 | ||
| α, β, γ cell (°) | 90, 90, 120 | 90, 90, 120 | ||
| Peak-1 | Inflection-1 | Remote-1 | ||
| Wavelength (Å) | 0.97949 | 1.71076 | 1.71145 | 1.70371 |
| Resolution (Å) | 30.9–1.35 | 31.1–2.7 | 31.1–2.7 | 31.1–2.2 |
| Rmerge | 0.046 (0.339) | 0.07 (0.30) | 0.07 (0.29) | 0.06 (0.88) |
| *CC1/2 | (0.839) | |||
| Mean I/σ(I) | 28.2 (3.1) | 33.7 (8.1) | 35.2 (8.0) | 31.6 (1.3) |
| Completeness (%) | 95.2 (68.6) | 100 (100) | 99.9 (99.0) | 99.0 (90.7) |
| Redundancy | 4.8 (3.0) | 18.5 (17) | 18.4 (16.7) | 17.0 (8.7) |
| Refinement | ||||
| Resolution (Å) | 30.8–1.35 | 31.1–2.2 | ||
| No reflections | 16,503 | 3,740 | ||
| Rwork/Rfree | 0.160 (0.214)/0.198 (0.250) | 0.227 (0.377)/0.275 (0.396) | ||
| No atoms | ||||
| Protein | 516 | 473 | ||
| Ligand/ion | 1 | 48 | ||
| Water | 75 | 31 | ||
| B-factors | ||||
| Protein | 27.1 | 34.0 | ||
| Ligand/ion | 17.9 | 68.0 | ||
| Water | 51.4 | 36.7 | ||
| R.m.s.d.'s | ||||
| Bond lengths (Å) | 0.009 | 0.010 | ||
| Bond angles (°) | 0.954 | 1.290 | ||
| Ramachandran plot | ||||
| Favoured (%) | 98.3 | 98.5 | ||
| Allowed (%) | 1.7 | 1.5 | ||
| Outliers (%) | 0 | 0 | ||
*Highest resolution shell is shown in parenthesis.
*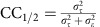 (ref. 61).
(ref. 61).
