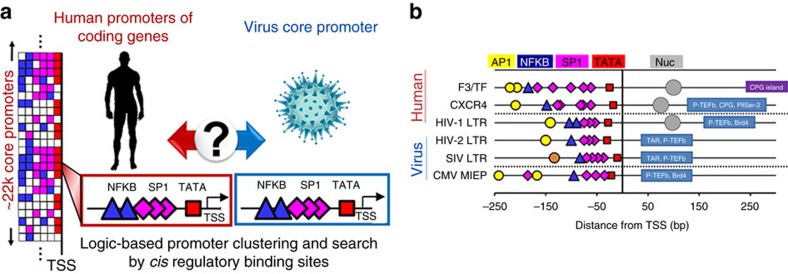
Figure 1. Viral and host promoters display high similarity in gene regulatory sequence.
(a) Logic-based search of human coding gene promoters for similarity to viral core promoters. The list of human promoters indicates a logic series of cis-binding elements (boxes) for each promoter (rows) comprising diverse patterns of TATA box (red), SP1 (pink) and NFKB (blue) binding sites. Conserved cis-binding sites in a virus (NFKB: blue triangles, SP1: pink diamonds and TATA: red squares) are searched for in all human coding genes. HIV and CMV promoters are curated and compared within ±250 bp from the TSS of human promoters. Promoters are clustered regardless of number, arrangement or distance of sites from the TSS. Additional transcription factor-binding site types are ignored to de-constrain results of the search. (b) The genome-wide promoter search results in ∼366 gene promoters with binding sites for TBP (TATA-binding protein, red squares), SP1 (pink diamonds) and NFKB (blue triangles), and F3/TF, CXCR4 and HIV-1 LTR show the highest similarity in cis regulatory arrangement. Upstream existence of AP1 sites (yellow circles) and downstream nucleosome occupancy (grey circles), binding of P-TEFb (ref. 69), BRD4 (ref. 69) and CPG islands (purple and navy rectangle regions) from the TSS are also consistent between promoters and curated after the initial genome-wide search. The brown circle in the SIV LTR promoter represents simian factor 1 or SF1, an activator protein 1 (AP1) related transcription factor in monkeys reported in SIV around −135 to −131 bp76.