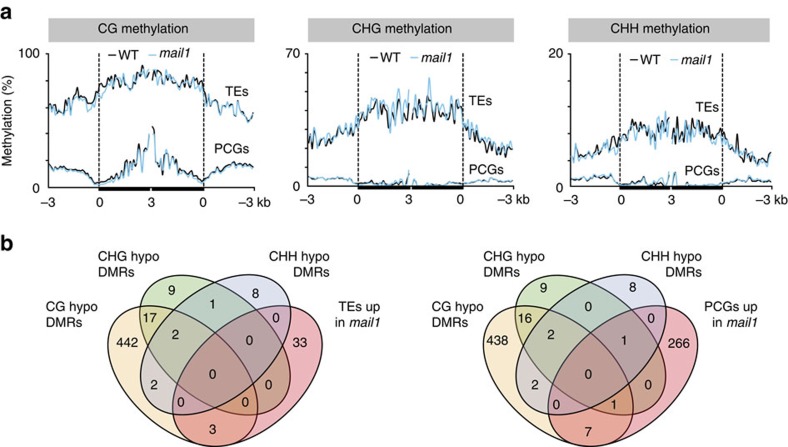
Figure 3. DNA methylation is not impaired in mail1.
(a) PCGs or TEs significantly upregulated in mail1 were aligned at the 5′-end (left dashed line) or the 3′-end (right dashed line), and average methylation levels in CG, CHG and CHH sequence contexts for each 100-bp bin in WT and mail1 were plotted from 3 kb upstream to 3 kb downstream. (b) Venn diagrams of overlap between genomic regions differentially hypomethylated in mail1 (hypo DMRs) in the three cytosine sequence contexts and TEs and PCGs significantly upregulated in mail1.