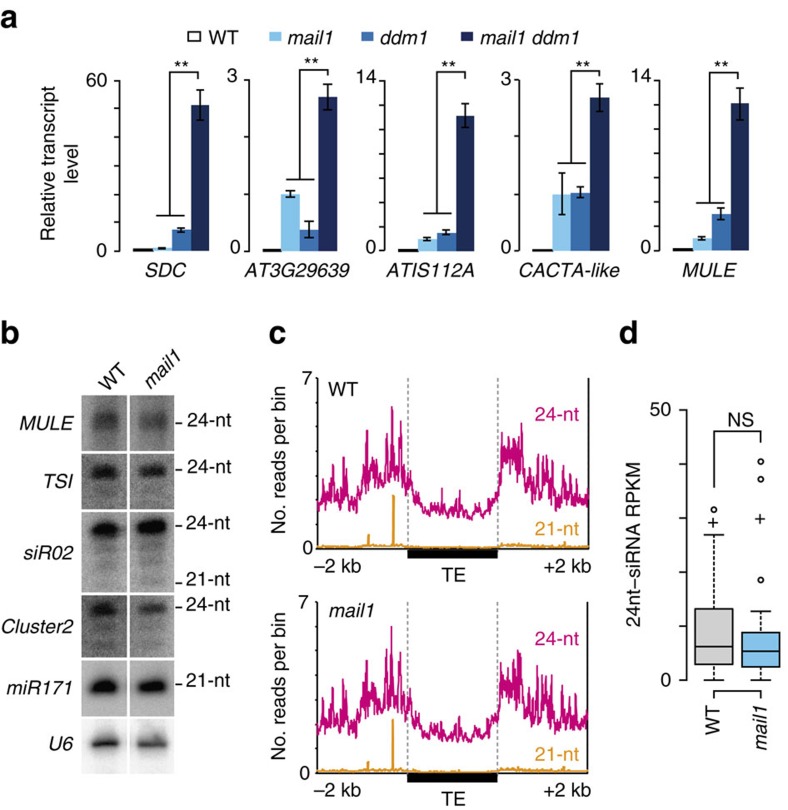
Figure 4. Loss of silencing in mail1 is independent of DNA methylation and siRNA accumulation.
(a) Expression analysis of mail1-overexpressed loci by quantitative RT–PCR in the indicated genotypes. mail1 levels are set to 1; values represent means from two to three biological replicates±s.e.m. Asterisks mark statistically significant differences from the mail1 ddm1 double mutant (unpaired, two-sided Student's t-test, P<0.05). (b) RNA gel blot analysis of siRNA accumulation. U6 RNA is shown as loading control. (c) 21- and 24-nt siRNA levels at TEs are unaltered in mail1. Number of uniquely mapped 21- and 24-nt siRNAs plotted along all Arabidopsis thaliana TEs from 2 kb upstream to 2 kb downstream of the TE annotation. Upstream and downstream sequences were divided each into 2,000 one-nucleotide bins, and TEs were divided into 2,000 bins of equal length. Average siRNA levels were computed for each bin in WT and mail1. (d) Boxplot of 24-nt siRNA accumulation level at mail1-upregulated TEs. Each box encloses the middle 50% of the distribution with the center lines marking the medians. Crosses mark sample means. Whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. Outliers are represented by circles. NS, nonsignificant (unpaired, two-sided Student's t-test, P>0.9).