Abstract
Bowen-Conradi syndrome (BCS) is a severe genetic disorder that is characterised by various developmental abnormalities, bone marrow failure and early infant death. This disease is caused by a single mutation leading to the aspartate 86 to glycine (D86G) exchange in the essential nucleolar RNA methyltransferase EMG1. EMG1 is required for the synthesis of the small ribosomal subunit and is involved in modification of the 18S ribosomal RNA. Here, we identify the pre-ribosomal factors NOP14, NOC4L and UTP14A as members of a nucleolar subcomplex that contains EMG1 and is required for its recruitment to nucleoli. The BCS mutation in EMG1 leads to reduced nucleolar localisation, accumulation of EMG1D86G in nuclear foci and its proteasome-dependent degradation. We further show that EMG1 can be imported into the nucleus by the importins (Imp) Impα/β or Impβ/7. Interestingly, in addition to its role in nuclear import, binding of the Impβ/7 heterodimer can prevent unspecific aggregation of both EMG1 and EMG1D86G on RNAs in vitro, indicating that the importins act as chaperones by binding to basic regions of the RNA methyltransferase. Our findings further indicate that in BCS, nuclear disassembly of the import complex and release of EMG1D86G lead to its nuclear aggregation and degradation, resulting in the reduced nucleolar recruitment of the RNA methyltransferase and defects in the biogenesis of the small ribosomal subunit.
Introduction
Ribosomes are large ribonucleoprotein (RNP) complexes that are required for the synthesis of all cellular proteins. The biogenesis of eukaryotic ribosomes is a highly dynamic and energy consuming cellular process that is closely coordinated with cell growth and proliferation (1–3). Ribosome production involves the assembly of approximately 80 ribosomal proteins as well as the transcription and processing of the ribosomal (r)RNA precursors and the chemical modification of specific residues in the rRNA sequences (4–6). The RNA modifications, which are introduced by a host of small nucleolar (sno)RNPs and lone-standing RNA methyltransferases, cluster in functionally important regions of the ribosome. This includes the decoding site, the intersubunit bridges and the peptidyl transferase centre, where the modifications function in stabilisation of the ribosome structure and can affect translation efficiency and fidelity (4,6,7). In humans, ribosome maturation requires the co-ordinated action of more than 200 biogenesis cofactors, including many enzymatic proteins such as exo- and endonucleases, RNA helicases, GTPases and RNA methyltransferases, which are thought to catalyse irreversible steps, thereby driving the directionality of the pathway (5,8–11). The pre-ribosomal recruitment of ribosomal proteins and biogenesis factors, which are translated in the cytoplasm and mostly imported into the nucleus and recruited to the nucleolus, is a tightly regulated and hierarchical process to ensure the correct assembly of the ribosomal subunits.
An increasing number of autosomal inherited human disorders have been linked to mutations in ribosomal proteins or factors involved in ribosome synthesis (12–14). Such diseases, which are termed ribosomopathies, are often characterised by developmental defects, craniofacial abnormalities and haematological dysfunction. Paradoxically, although the tumour suppressor p53 is stabilised when ribosome synthesis is perturbed and is therefore dysregulated in many ribosomopathies, these diseases often pre-dispose patients to cancer (12,15,16). The emerging family of ribosomopathies includes Diamond Blackfan anaemia (OMIM 105650), a severe bone marrow failure syndrome that is linked to mutations in various ribosomal proteins (17,18) and the craniofacial disorder Treacher Collins syndrome (OMIM 154500/613717), which is caused by mutations in various components of RNA polymerase I (19,20). Similarly, mutations in NOL11, a ribosome biogenesis factor that is required for pre-rRNA transcription and processing, lead to North American Indian childhood cirrhosis (OMIM 604901; 21,22). Shwachman-Bodian-Diamond syndrome (OMIM 260400), which manifests in bone marrow failure, skeletal abnormalities and a predisposition to leukaemia, is caused by a mutation in the late large subunit biogenesis factor SBDS that inhibits GTP hydrolysis by EFL1, preventing eIF6 release and thus impeding translation (23,24). Despite the identification of disease mutations in ribosomal proteins and ribosome biogenesis factors, many of these factors remain uncharacterised in human cells, and often an understanding of the molecular basis of these diseases is lacking.
Bowen-Conradi syndrome (BCS; OMIM 211180) is a severe autosomal recessive disorder characterised by pre- and postnatal psychomotor defects, growth retardation, microcephaly, micrognathia and rocker bottom feet, and leads to early infant death (25–27). Interestingly, BCS was recently shown to be caused by a single amino acid exchange (aspartate to glycine) at position 86 of the nucleolar RNA methyltransferase EMG1 (28). The yeast orthologue of EMG1 (Emg1, also known as Nep1 (29)), is required for biogenesis of the small ribosomal subunit, where it participates in the hypermodification of uridine 1191 of the 18S rRNA (30–32). Consistent with this, Emg1 was identified to bind close to this site in a recent cryo-EM structure of a 90S pre-ribosome from Chaetomium thermophilum (33). Uridine 1191 is first isomerised to pseudouridine by the box H/ACA snoRNP snR35, which generates the substrate for N1 methylation by Emg1. Subsequently, a 3-amino-3-carboxypropyl (acp) modification is added in the cytoplasm by Tsr3 to generate the N1-methyl-N3-aminocarboxypropylpseudouridine (m1acp3Ψ) modification (34). However, similar to other pre-rRNA methyltransferases, such as Bud23/WBSCR22, the catalytic activity of Emg1 is not essential for ribosome biogenesis, but rather the presence of the protein within the pre-ribosomal complex is required (32,35,36). Structural analyses of Emg1 (35,37,38) indicated that aspartate 86 normally forms a salt-bridge with arginine 84 and it has been suggested that the glycine substitution found in BCS disrupts this interaction, leading to destabilisation of the protein (28). Consistent with this, EMG1 levels were found to be reduced in BCS (28), but how this mutation might influence EMG1 function and the molecular basis of BCS has remained unknown.
Here we show that EMG1 carrying the BCS mutation is destabilised and mislocalises to the nucleoplasm where it accumulates in nuclear foci. Our data demonstrate that both wild-type EMG1 and BCS mutant EMG1 can be recruited to the nucleolus by a subcomplex composed of NOP14, NOC4L and UTP14A. We further show that while EMG1 can also be imported into the nucleus by the classical importin α/importin β (Impα/β) pathway, it is an import substrate of the importin β/importin 7 (Impβ/7) heterodimer, which is not only responsible for nuclear import of EMG1 but also stabilises EMG1 by functioning as a chaperone. Together, our data suggest that in BCS EMG1D86G is chaperoned by Impβ/7 in the cytoplasm and during nuclear import, but once released, it partially aggregates in the nucleus leading to the formation of nuclear foci and to proteasome-dependent degradation. Although a fraction of EMG1D86G can still be recruited to the nucleolus, decreased nucleolar EMG1 levels lead to defects in the biogenesis of the small ribosomal subunit.
Results
The BCS mutation causes accumulation of EMG1D86G in nuclear foci and degradation of the protein
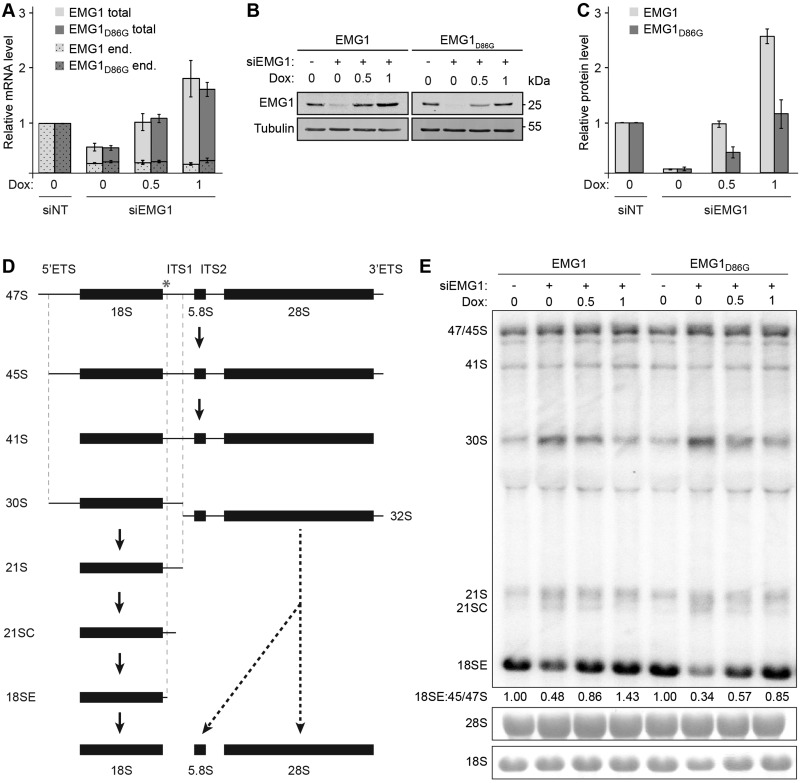
The RNA methyltransferase Emg1, which is mutated in BCS, has been shown to participate in the hypermodification of uridine 1191 in the yeast 18S rRNA. The catalytic activity of yeast Emg1 was found not to be essential, while the presence of the protein is required for biogenesis of the small ribosomal subunit (30–32,35). To allow analysis of the localisation, nucleolar targeting and function of EMG1 in human cells and to reveal the effects of the BCS mutation, we first established an RNAi-based rescue system in which endogenous EMG1 could be depleted and expression of untagged, siRNA-resistant wild-type EMG1 or EMG1 carrying the BCS mutation (EMG1D86G) could be induced by addition of doxycycline. Treatment of these cells with siRNAs targeting EMG1 depleted the endogenous EMG1 mRNA and addition of doxycycline allowed re-expression of the wild-type EMG1 or EMG1D86G mRNAs to similar levels at each of the doxycycline concentrations used (Fig. 1A). Analysis of EMG1 protein in the corresponding samples using an antibody that detects both the endogenous and tet-induced EMG1, revealed that the EMG1 protein level was reduced to <7% when expression of the EMG1 transgenes was not induced (Figs. 1B and C). Importantly, while addition of 0.5 ng/ml doxycycline induced expression of wild-type EMG1 to the same level as the endogenous protein, the level of EMG1D86G was more than two-fold lower (Fig. 1B and C). Similarly, the addition of 1 ng/ml doxycycline induced expression of EMG1D86G similar to endogenous levels, while the same concentration of doxycycline caused significant over-expression of the wild-type protein, implying that the BCS mutation destabilises the EMG1 protein. This finding is in line with previous observations showing decreased EMG1 levels in BCS patient cells (28). Furthermore, and consistent with this model, treatment of cells expressing EMG1D86G with the proteasome inhibitor MG132 resulted in stabilisation and an accumulation of the mutant protein, implying that EMG1D86G is largely degraded in a proteasome-dependent manner (Supplementary Material, Fig. S1).
Figure 1.
The Bowen-Conradi syndrome mutation destabilises the EMG1 protein. (A) HEK293 cell lines were transfected with non-target siRNAs (siNT) or siRNAs against EMG1 (siEMG1) and RNAi was performed for 72 h. 24 h before harvesting, expression of untagged wild-type EMG1 (EMG1) or EMG1 carrying the Bowen-Conradi syndrome mutation (EMG1D86G) was induced by addition of doxycycline at the concentrations indicated (ng/ml). After harvesting of cells, RNA was extracted and the level of the endogenous EMG1 mRNA (end.), and the endogenous and tet-induced EMG1 mRNAs (total), normalised to the GAPDH mRNA was determined by qPCR. Data from three independent experiments are shown as mean +/- SEM. (B-C) Proteins extracted from siRNA-treated cells prepared as described in (A) were analysed by SDS-PAGE followed by western blotting using antibodies against EMG1 and Tubulin (B), and the levels of EMG1 protein, normalised to Tubulin were quantified (C). Data from three independent experiments are shown as mean +/- SEM. (D) Simplified schematic overview of pre-rRNA processing in human cells. Mature rRNAs are shown as filled rectangles and spacers as lines. ETS – external transcribed spacer; ITS – internal transcribed spacer. An asterisk indicates the target site on the 47S pre-rRNA of the probe used for northern blotting. (E) Pre-rRNA levels in total cellular RNA prepared as described in (A) were analysed by northern blotting using a probe hybridising to the 5’ end of ITS1. Numbers below the northern blot panel indicate the relative quantitation of the 18SE pre-rRNA compared to 47/45S pre-rRNAs. The mature 18S and 28S rRNAs were visualised by methylene blue staining.
Analysis of precursor rRNA levels in these cells by northern blotting showed that siRNA-mediated depletion of EMG1 causes accumulation of the 30S, 21S and 21SC pre-rRNAs and leads to a decrease in 18SE pre-rRNA levels (Figs. 1D and E), confirming the role of human EMG1 in the biogenesis of the 18S rRNA. Interestingly, these pre-rRNA processing defects could be rescued by expression of either wild-type EMG1 or EMG1D86G (Fig. 1E) at the endogenous protein level (Fig. 1B and C; 0.5 and 1 ng/ml doxycycline for EMG1 and EMG1D86G, respectively). These results indicate that the EMG1 mutation in BCS does not directly affect the function of the protein in ribosome biogenesis, but that the decreased level of the EMG1D86G protein leads to impaired maturation of the small ribosomal subunit.
Having established a cell system that mimics the BCS phenotype, we next analysed the localisation of untagged wild-type and mutant EMG1 by immunofluorescence to gain further insight into the molecular basis of the disease. Similar to the yeast protein, wild-type EMG1 primarily localises to the nucleolus in human cells. In contrast, although EMG1D86G was also partially localised in the nucleolus and no significant change in nucleolar structure was observed, a large fraction of the protein was detected in the nucleoplasm and in particular, was found to accumulate in nuclear foci (Fig. 2), indicating that the BCS mutation in EMG1 can lead to its mislocalisation. Together, these data demonstrate that the EMG1D86G protein is destabilised and accumulates in nuclear foci, leading to its proteasome-dependent degradation, which is in line with the reduced protein levels reported for BCS patient cells. Furthermore, the decreased amount of EMG1D86G in the nucleolus leads to defects in the production of the small ribosomal subunit and only when EMG1D86G is overexpressed is it able to fulfil its role in 18S rRNA maturation.
Figure 2.
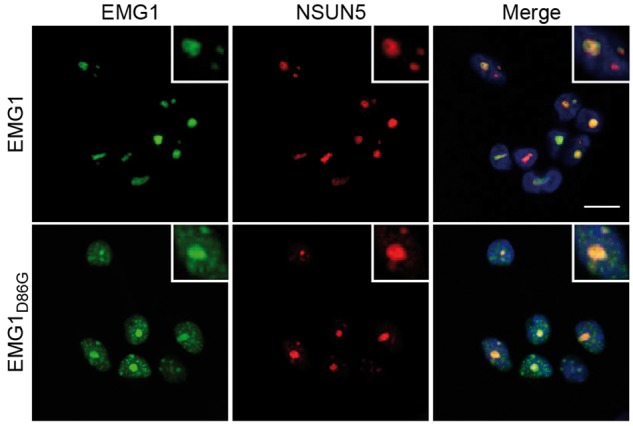
EMG1D86G accumulates in nuclear foci HEK293 cell lines were transfected with siRNAs against EMG1 and the expression of untagged, siRNA-insensitive wild-type EMG1 (EMG1) or EMG1 carrying the Bowen-Conradi syndrome mutation (EMG1D86G) was induced at the endogenous level by the addition of doxycycline. Cells were fixed and immunofluorescence was performed using antibodies against EMG1 (green) and NSUN5 (nucleolar marker; red), and nuclei were visualised by DAPI staining (blue) in the overlay (merge). Scale bar indicates 10 μm.
Nucleolar recruitment of wild-type EMG1 and the BCS mutant requires NOP14, NOC4L and UTP14A
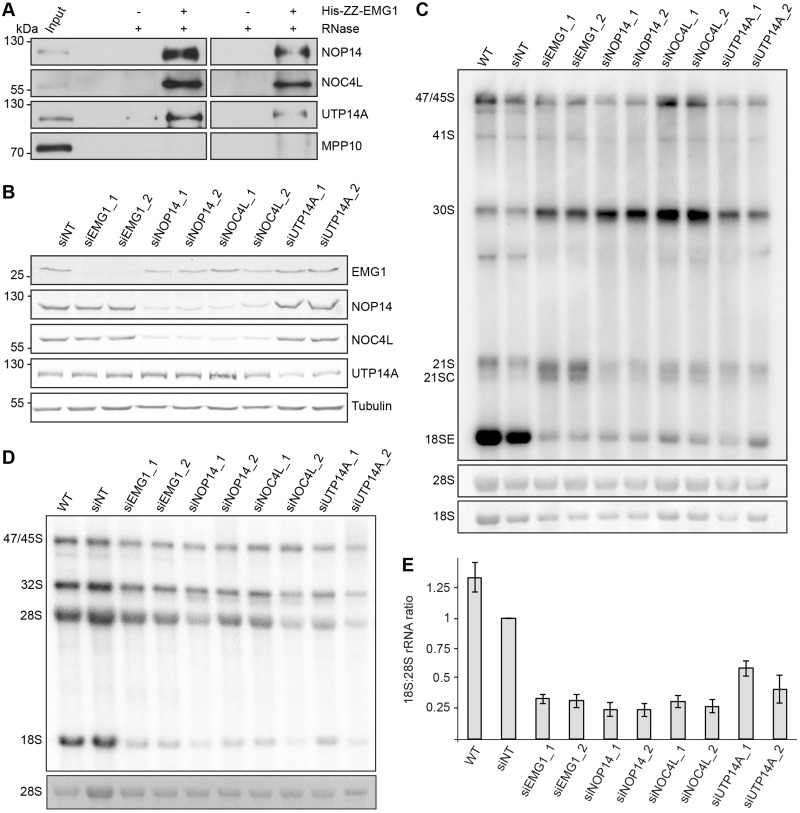
The finding that EMG1D86G accumulates in the nucleoplasm and in nuclear foci, raised the possibility that the BCS mutation prevents specific recruitment of EMG1 to the nucleolus, which might then lead to its destabilisation. To understand how EMG1 is normally recruited to its specific binding sites on nucleolar pre-ribosomes, we performed immunoprecipitation experiments to identify interaction partners of EMG1. HEK293 cells expressing Flag-tagged EMG1, or the Flag-tag alone as a control, were used to immunoprecipitate EMG1-containing complexes. Analysis of co-precipitated proteins by mass spectrometry identified NOP14 and NOC4L as significantly enriched with EMG1 compared to the control sample, suggesting that these interactions are conserved from yeast to humans (30,39). In addition, however, UTP14A was observed to specifically co-precipitate with EMG1, implying that this factor may also contribute to the function of EMG1 in human cells. To confirm these interactions, binding assays were performed with recombinant His-ZZ-tagged EMG1 immobilised on IgG sepharose, which was incubated with HeLa cell extract. Proteins retained with EMG1 were then detected by western blotting (Fig. 3A), which demonstrated that NOP14, NOC4L and UTP14A, but not another pre-SSU component MPP10, specifically bind to EMG1. Furthermore, RNase treatment of the cell extract did not affect these interactions, suggesting that these proteins likely form a subcomplex via protein-protein interactions that are not mediated via RNA or pre-ribosomes.
Figure 3.
EMG1 is part of a pre-ribosomal subcomplex containing NOP14, NOC4L and UTP14a. (A) His-ZZ-tagged EMG1 was immobilised on IgG sepharose and after incubation with HeLa cell lysate, complexes were treated with RNase (+) or left untreated (-). EMG1 and co-precipitated proteins were eluted by cleavage using TEV protease and eluates were analysed by western blotting using antibodies against the indicated endogenous proteins. (B) HeLa cells were transfected with non-target siRNAs (siNT) or siRNAs against NOP14, NOC4L, UTP14A or EMG1 and harvested after 72 h. Proteins were analysed by western blotting using antibodies against the indicated ribosome biogenesis cofactors, and tubulin as loading control. (C) HeLa cells were treated with siRNAs as in (B), total RNA was extracted and pre-rRNA levels were monitored by northern blotting using a probe hybridising the 5’ end of ITS1. The mature 18S and 28S rRNA, visualised by methylene blue staining, served as a loading control. (D) HeLa cells were transfected with siRNAs as in (B) and after 72 h, cells were pulse-labelled with 32P-orthophosphate and then grown in normal media for a further 3 h. Total RNA was extracted, separated by agarose-glyoxal gel electrophoresis and newly synthesised precursor and mature rRNAs were detected using a phosphorimager. Mature 28S rRNA, visualised by ethidium bromide staining, served as a loading control. (E) The ratio of newly synthesised 18S to 28S rRNAs was quantified in the pulse-labelling experiments and data from three independent experiments are shown as mean +/- SEM.
To further investigate whether EMG1, NOP14, NOC4L and UTP14 function together, we examined the effects of individual depletion of each of these proteins on ribosome maturation. RNAi-mediated depletion of these factors was established (Fig. 3B) and, interestingly, it was observed that depletion of NOP14 led to a decrease in NOC4L levels and vice versa, suggesting that the levels of these proteins are co-regulated (Fig. 3B). Furthermore, depletion of any of the four proteins led to an accumulation of the 30S pre-rRNA and a concomitant decrease in 18SE pre-rRNA and mature 18S rRNA levels (Fig. 3C–E), supporting the model that these proteins act together in the biogenesis of the small ribosomal subunit.
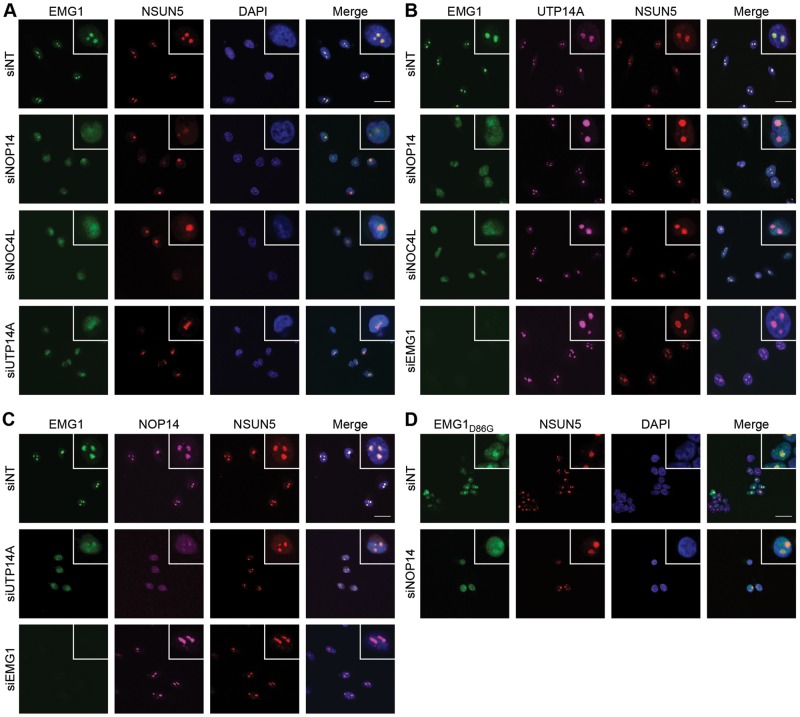
We therefore examined the effect of depleting individual members of this subcomplex on the localisation of the other components. Interestingly, depletion of either NOP14, NOC4L or UTP14A led to mislocalisation of EMG1 to the nucleoplasm, indicating that these three proteins are all required for recruitment of EMG1 to the nucleolus (Fig. 4A). NSUN5 served as a nucleolar marker and was retained in the nucleolus upon knockdown of NOP14, NOC4L or UTP14A, showing the specificity of their role in the nucleolar recruitment of EMG1. In contrast, depletion of NOP14, NOC4L or EMG1 had no effect on the nucleolar localisation of UTP14A (Fig. 4B). Furthermore, depletion of UTP14A, but not EMG1, caused mislocalisation of NOP14 to the nucleoplasm (Fig. 4C), suggesting a hierarchical mode of recruitment in which the presence of UTP14A is a prerequisite for the nucleolar association of all other members of the subcomplex and where NOP14 and NOC4L are also required for recruitment of EMG1.
Figure 4.
Hierarchical recruitment of EMG1, NOP14/NOC4L and UTP14A into the nucleolus. (A–C) HeLa cells were transfected with non-target siRNAs (siNT) or those targeting the factors indicated to the left of each panel and the localisation of EMG1 (green), NSUN5 (red) and UTP14A or NOP14 (purple) was determined by immunofluorescence using antibodies against the endogenous proteins. Nuclear material was visualised by DAPI staining (blue). The scale bar indicates 20 μm. (D) HEK293 cell lines expressing siRNA-resistant, untagged EMG1 or EMG1D86G at endogenous levels were transfected with siRNAs against endogenous EMG1 and co-transfected with either non-target siRNAs (siNT) or siRNAs against NOP14. The localisation of EMG1 and EMG1D86G was determined by immunofluorescence using an anti-EMG1 antibody. Nuclear material was visualised by DAPI staining (blue). The scale bar indicates 20 μm.
Having identified the nucleolar interaction partners of EMG1, we next investigated whether the partial nucleolar localisation of EMG1D86G is due to specific recruitment by these interaction partners or whether the nucleolar fraction of EMG1D86G accumulates non-specifically in the nucleolus. Cells depleted of endogenous EMG1 and expressing untagged, siRNA-resistant EMG1D86G to the endogenous levels were treated with non-target siRNAs or siRNAs against NOP14 before visualisation of EMG1D86G and NSUN5 by immunofluorescence (Fig. 4D). Similar to the mislocalisation of wild-type EMG1 protein observed upon depletion of its nucleolar recruitment factors, depletion of NOP14 caused loss of EMG1D86G from the nucleolus, implying that the mutant protein is able to interact with its partners and can be specifically recruited to its pre-ribosomal binding sites in the nucleolus. Consistent with this and the observation that only a fraction of EMG1D86G is recruited to the nucleolus, NOC4L, NOP14 and UTP14A were specifically co-immunoprecipitated with Flag-EMG1D86G, albeit at slightly lower levels than with the wild-type protein (Supplementary Material, Fig. S2A). To analyse whether the co-immunoprecipitation of the other members of the subcomplex might also be due to their aggregation together with EMG1D86G, we investigated whether these proteins co-localise with the BCS mutant in the nucleoplasmic foci observed upon expression of EMG1D86G (Fig. 2). However, in cells expressing EMG1D86G, analysis of the localisation of NOP14 revealed that it remains predominantly in nucleoli and is not detected in nucleoplasmic foci together with EMG1D86G (Supplementary Material, Fig. S2B), indicating that the nucleoplasmic foci are specific to the BCS mutant and do not contain its interaction partners or pre-ribosomal complexes.
The finding that a portion of EMG1D86G can be recruited to the nucleolus (Fig. 4D), raises the question of whether the D86G mutation found in BCS affects the catalytic activity of the protein on the human 18S rRNA. We therefore performed in vitro methylation assays using recombinantly expressed EMG1 or EMG1D86G and RNA oligonucleotides containing fragments of the 18S rRNA sequence (1245–1255) that span the EMG1 target residue (U1248 or Ψ1248) in the presence of 3H-labelled S-adenosylmethionine. This confirmed that, similar to the yeast protein (32), both EMG1 and EMG1D86G can methylate this position, but that methylation by EMG1D86G is less efficient and that the isomerisation of the uridine to pseudouridine is a pre-requisite for N1-methylation (Supplementary Material, Fig. S2C).
Nuclear import of EMG1 can be mediated by Impβ/7 or Impα/β
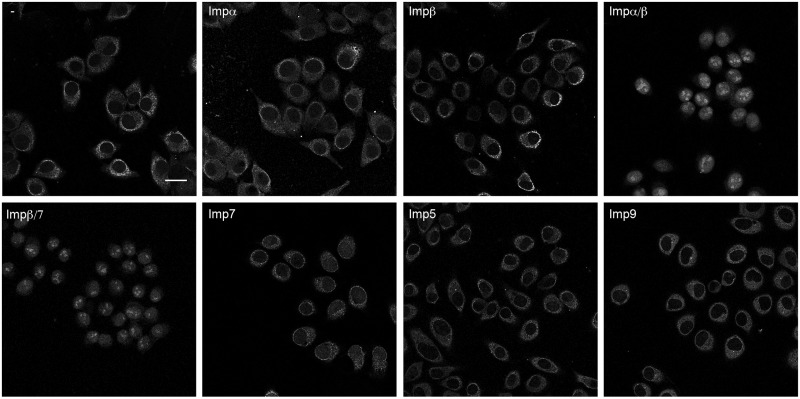
In line with the reduced EMG1 protein levels observed in BCS patients, our data show that the D86G mutation decreases the stability of the EMG1 protein and leads to the formation of speckles that likely represent protein aggregates. Interestingly, these foci are observed to form in the nucleoplasm impeding the nucleolar recruitment of a large fraction of the protein. This raises the question of how EMG1 reaches the nucleoplasm after its translation in the cytoplasm and we therefore aimed to identify the import receptor(s) responsible for nuclear import of EMG1. Binding assays revealed specific association of Impα, Impβ and Imp7 with EMG1 only in the absence of RanGTP, suggesting that their interaction occurs under cytoplasmic conditions (where RanGTP levels are very low) and in a RanGTP-regulated manner, and that these factors might therefore represent import receptors for EMG1. Import assays were performed using permeabilised HeLa cells together with recombinantly expressed importins and green fluorescent protein (GFP)-tagged EMG1 in the presence of a reconstituted RanGTP gradient. Permeabilisation of the cell membrane with a low concentration of digitonin releases the cytoplasm, enabling endogenous importins to be removed, while leaving the nuclear membrane intact. Incubation of GFP-EMG1 with permeabilised cells in the absence of exogenous importins or in the presence of Imp5, Imp7, Imp9, Impα or Impβ individually did not enable nuclear import of EMG1 (Fig. 5). In contrast, addition of either Impα together with Impβ or the Impβ/7 heterodimer lead to efficient nuclear import of GFP-EMG1, indicating that EMG1 can be a substrate of both the Impα/β and Impβ/7 import pathways (Fig. 5).
Figure 5.
Nuclear import of Emg1 can be mediated by Impα/β or the Impβ/7 heterodimer. Import of GFP-tagged EMG1 into the nuclei of permeabilised HeLa cells was performed in the presence of Ran, an energy-regenerating system and the importins indicated. The localisation of EMG1 was determined by confocal fluorescence microscopy. The scale bar indicates 25 μm.
The importin β/7 heterodimer can prevent precipitation of EMG1 and EMG1D86G
Besides their primary role in the nuclear import of proteins and macromolecular complexes, several importins have been shown to act as chaperones by preventing aggregation of basic proteins on polyanions in the cytoplasm (Fig. 6A; 40). A prominent example among these is the Impβ/7 heterodimer, which plays an essential role in maintaining the solubility of highly basic proteins, such as the ribosomal proteins RPL4 (uL4) and RPL6 (eL6) and the histone H1 (40–42). Structural analysis of Emg1 has revealed a highly conserved basic patch on the surface of EMG1 that likely mediates RNA binding and within which the D86G mutation is found (35,37). We therefore investigated whether Impβ/7 could also serve as a chaperone for EMG1 and EMG1D86G. Recombinantly expressed, soluble EMG1 was incubated with polyanions (transfer (t) RNA) and after separation of the soluble and insoluble fractions by centrifugation, EMG1 was found to be largely precipitated (Fig. 6B). However, the addition of Impβ/7, but not bovine serum albumin (BSA), prior to tRNA addition and centrifugation led to EMG1 remaining in the soluble fraction (Fig. 6B), indicating that the importin complex binds to EMG1 and prevents its nonspecific interaction and precipitation with the tRNA. Since Impα/β can also bind to EMG1 and facilitate its nuclear import, we also tested whether the Impα/β complex could function as a chaperone for EMG1. Although addition of Impα/β also decreased the precipitation of EMG1, this only occurred to a lesser extent than what was observed for Impβ/7 (Supplementary Material, Fig. S3). Due to the inherent instability of EMG1D86G, analysis under similar conditions led to the precipitation of the mutant protein even in the absence of tRNA. However, performing the assay at a lower pH (pH 7.1) where EMG1D86G is less basic and therefore more soluble revealed that while EMG1D86G is mostly precipitated upon addition of tRNA alone, it largely remains soluble in the presence of Impβ/7 (Fig. 6C). Together, these results indicate that the Impβ/7 heterodimer acts as a chaperone preventing precipitation of EMG1/EMG1D86G in the cytoplasm, explaining why EMG1D86G is efficiently imported, then aggregates in nuclear foci and is degraded in BCS.
Figure 6.
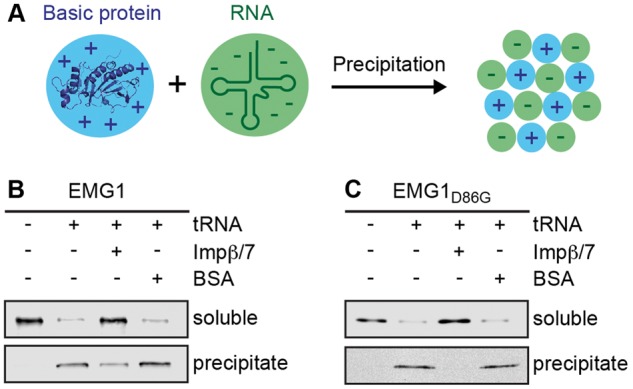
The importinβ/7 complex suppresses EMG1 precipitation in the presence of polyanions. (A) Schematic view of the precipitation of basic proteins in the presence of polyanions such as nucleic acids. (B) Soluble His-ZZ-tagged EMG1 alone or together with recombinantly expressed Impβ/7 or BSA was incubated with tRNA. After centrifugation, soluble and precipitated proteins were analysed by western blotting using an antibody against EMG1. (C) The anti-aggregation assay was performed using EMG1D86G as described in (B), but under less basic conditions to improve solubility of the mutant protein.
Discussion
Bowen-Conradi syndrome (BCS; OMIM 211180) is a member of the growing family of genetic diseases termed ribosomopathies that are caused by mutations in genes coding for ribosomal proteins or ribosome biogenesis cofactors (12–14). Many of these defects inhibit biogenesis of the small ribosomal subunit and they often predominantly affect rapidly dividing cells, leading to haematological disorders and craniofacial defects. In the case of BCS, a single point mutation in the gene encoding the RNA methyltransferase EMG1 leads to various developmental defects including pre- and post-natal growth retardation, joint abnormalities, microcephaly, micrognathia and rocker bottom feet (25,28). Due to the severity of the disorder, most of the BCS patients do not survive the first year of their life. Analysis of patient material (28,43), a knockin mouse model (44) and the results presented here show that the aspartate 86 to glycine mutation in EMG1 observed in BCS causes mislocalisation and degradation of the protein leading to defects in biogenesis of the small ribosomal subunit. Based on the findings that the catalytic activity of yeast Emg1 is not required for ribosome synthesis and that introduction of the BCS mutation in the Methanocaldococcus jannashii EMG1 homologue (45), yeast Emg1 (32) or human EMG1 (Supplementary Material, Fig. S2C) only mildly affected its catalytic activity, it is likely that the BCS is caused by the absence of EMG1 from its pre-ribosomal binding sites in the nucleolus, rather than by a lack of the rRNA modification. Furthermore, the observations that the EMG1D86G protein is present at lower cellular levels than wild-type EMG1 and that when EMG1D86G is overexpressed it is able to fulfil its functions in the maturation of the small ribosomal subunit, strongly suggest that the decreased EMG1 levels caused by the D86G mutation are the basis of BCS.
We have analysed the nuclear import of EMG1 from its sites of translation in the cytoplasm and describe its nucleolar recruitment by specific interaction partners (Fig. 7; see scheme on the left). EMG1 can be specifically imported into the nucleus by Impα/β or by the Impβ/7 heterodimer. Import complexes disassemble in the nucleus upon binding of RanGTP to the importins and EMG1 establishes interactions with NOP14, NOC4L and UTP14A that facilitate its nucleolar recruitment, enabling the RNA methyltransferase to fulfil its function in the biogenesis of the small ribosomal subunit. Interestingly, the heterodimer of Imp7 and Impβ not only mediates the translocation of EMG1 into the nucleus, but also acts as a chaperone that prevents aggregation of EMG1 on cellular RNAs, most likely by binding to the basic patches on the RNA methyltransferase. This role of the nuclear import machinery has been observed for a variety of basic nuclear proteins, including several ribosomal proteins and histones (40–42). In the case of EMG1, import pathways constituted by both Impα/β or Impβ/7 can mediate translocation of EMG1 into the nucleus, however, only the Impβ/7 complex can efficiently prevent EMG1 aggregation on RNAs, suggesting that this complex represents the physiological import receptor. Interestingly, some nuclear proteins, such as the ribosomal proteins Rpl4, Rpl5 and Rpl11, require specific chaperones that bind during nuclear import but do not play a direct role in translocation, and might allow further levels of regulation for these pathways (46–48). For EMG1, the Impβ/7 complex is sufficient to mediate nuclear import without additional proteins, further suggesting that this represents a constitutive pathway that is essential for ribosome assembly in all cells. Remarkably, while most importin-cargo complexes are thought to disassemble readily upon interaction with nuclear RanGTP, Imp7-containing import complexes have been shown to be more stable in the presence of RanGTP than complexes of other importins (41,49). This raises the possibility that Imp7 might remain associated longer with its cargos in the nucleus, allowing basic proteins the time to bind to their specific interaction partners and thereby preventing unspecific interactions on nucleic acids.
Figure 7.
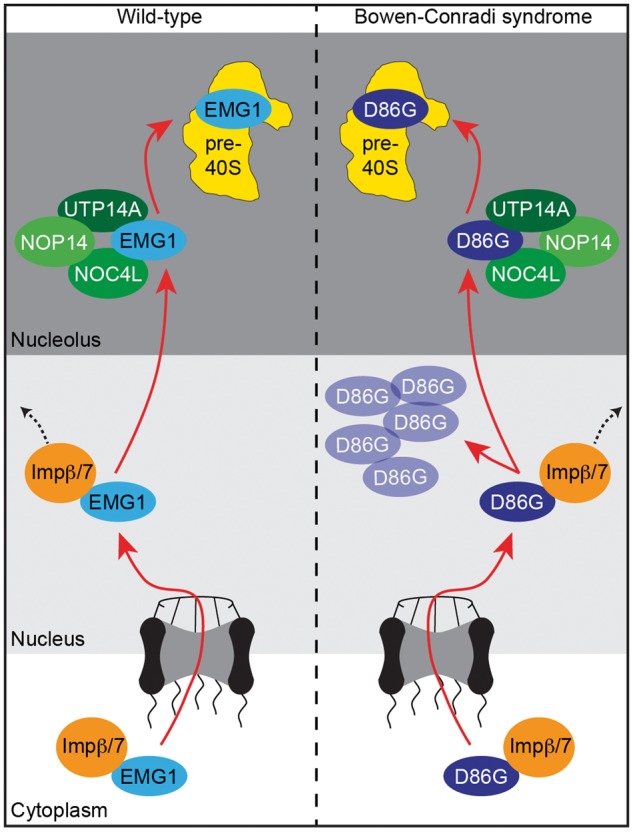
EMG1 in Bowen-Conradi syndrome. The Impβ/7 dimer binds to EMG1 in the cytoplasm and acts as a chaperone during nuclear import of EMG1. EMG1 is then recruited by a subcomplex containing NOP14, NOC4L and UTP14a to the nucleolus where is fulfils its essential functions in the biogenesis of the small ribosomal subunit. In Bowen-Conradi syndrome, EMG1D86G is chaperoned into the nucleus by Impβ/7, where it accumulates in nuclear speckles leading to proteasome-dependent protein degradation. Only a fraction of EMG1D86G can be recruited to the nucleolus and the decreased pre-ribosomal levels of EMG1D86G lead to defects in small ribosomal subunit maturation.
In BCS, the accumulation of EMG1D86G in nuclear foci implies that the binding of the importins prevents aggregation of the EMG1 mutant in the cytoplasm and during nuclear import, but that the nuclear disassembly of the import complex might then cause EMG1D86G to aggregate in the nucleoplasm (Fig. 7; see scheme of BCS on the right). Specifically for the BCS mutant, its residence in complex with the importins may not be sufficient to allow the formation of specific interactions with nucleolar factors. In line with this, we observed that only a fraction of EMG1D86G localises to nucleoli, where it can bind to its specific interaction partners. The majority of EMG1D86G accumulates in speckles in the nucleoplasm and is degraded in a proteasome-dependent manner, leading to the reduced EMG1 levels observed in BCS patient cells. Together, these insights into the dysfunction of EMG1D86G in BCS highlight the possibility of development of drugs that stabilise EMG1D86G for treatment of BCS.
Materials and Methods
Expression and purification of recombinant proteins
The EMG1 coding sequence (NM_006331.7) was cloned into a pQE80-derivative vector for recombinant expression of a His-ZZ- or His-GFP-fusion protein. Site-directed mutagenesis was used to introduce a point mutation to convert aspartate 86 to glycine as in BCS (28). Protein expression was induced in Escherichia coli BL21 Rosetta 2 by the addition of 250 μM isopropyl β-D-1-thiogalactopyranoside (IPTG) and cultures were grown overnight at 18 °C. Cells were lysed by sonication in a buffer containing 30 mM potassium phosphate (KPi) pH 8, 300 mM KCl, 10% (v/v) glycerol, 10 mM imidazole, 0.1 mM dithiothreitol (DTT) and protease inhibitor cocktail (cOmplete; Roche) and soluble lysates were then incubated with cOmplete His-tag Purification Resin (Roche), the matrix was washed with sonication buffer and proteins were eluted in buffer containing 30 mM potassium phosphate (KPi) pH 8, 300 mM KCl, 10% (v/v) glycerol, 500 mM imidazole and then dialysed into a buffer composed of 30 mM KPi pH 8, 100 mM KCl, 20% (v/v) glycerol, 0.1 mM ethylenediaminetetraacetic acid (EDTA), 0.1 mM DTT.
The coding sequences of Impα, Impβ, Imp5, Imp7 and Imp9 were cloned into pQE80-derivative vectors for recombinant expression (40) of His14-brSUMO or His14-brNEDD8-, or ZZ-brSUMO-tagged proteins. Expression and purification of proteins was performed as previously described (50). In brief, protein expression was induced in NEB express competent E. coli cells (New England Biosciences) grown overnight at 27 °C by the addition of 100 µM IPTG at 18 °C for 3 h. Cell lysis was achieved using sonication in Lysis buffer (50 mM Tris-HCl pH 7.5, 300 mM NaCl, 20 mM imidazole and 5 mM DTT). Tagged proteins in soluble lysates were immobilised on Ni-EDTA resin and the matrix was washed using Lysis buffer and a high-salt buffer containing 50 mM Tris-HCl pH 7.5, 2 M NaCl, 20 mM imidazole and 5 mM DTT. Elution was performed ‘on-column’ by protease cleavage in Lysis buffer supplemented with 250 mM sucrose. To produce the Impβ/7 heterodimer, Imp7 and Impβ were co-expressed and the heterodimer was purified as above followed by an additional purification step on antiZZ-affinity resin and protease cleavage elution.
Anti-aggregation assays using recombinant EMG1
Anti-aggregation assays for EMG1 were performed in 50 mM Tris-HCl pH 7.4 and 100 mM NaCl, as described previously for other basic proteins (40). As the EMG1D86G is inherently unstable under these conditions, assays were instead performed in 50 mM Tris-HCl pH 7.1 and 100 mM NaCl. Recombinant His-ZZ-tagged EMG1 or EMG1D86G (0.5 μM) was incubated with E.coli tRNA (30 μg/ml) in a reaction volume of 50 μl for 15 min at room temperature. Alternatively, before addition of tRNA, Impβ/7, Impα/β or BSA as a control, was then added to a final concentration of 0.75 μM and samples were incubated for 15 min at room temperature. Samples were centrifuged at 20,000 x g for 1 h and the soluble and insoluble fractions were analysed by SDS-PAGE and western blot using an antibody against EMG1.
Import assays
Preparation of permeabilised HeLa cells and import assays were performed as previously described (49) unless otherwise stated. In brief, cells were permeabilised on ice using 25 μg/ml digitonin for 7 min. Import reactions were performed using 0.75 μM GFP-tagged EMG1 and 1.5 μM import receptor as indicated in a buffer containing 20 mM 4-(2-Hydroxyethyl)-1-piperazine ethanesulfonic acid (HEPES) pH 7.0, 110 mM KOAc, 3.5 mM MgOAc2, 1 mM EGTA and 250 mM sucrose for 10 min at room temperature. An energy-regeneration system containing 0.5 mM ATP, 0.5 mM GTP, 10 mM creatine phosphate, 50 µg/ml creatine kinase and 5 U/ml NDP-kinase and Ran-mix containing of 3 μM RanGDP, 0.3 μM NTF2, 0.2 μM RanBP1 and 0.2 μM Rna1p were added to the reaction.
Identification of EMG1-interacting proteins
HEK293 stable cell lines were treated with 1 μg/ml doxycycline for 24 h to induce expression of Flag-tagged EMG1 or the Flag-tag alone. Cells were harvested and lysed by sonication in a buffer containing 50 mM Tris pH 7.4, 150 mM NaCl, 0.5 mM EDTA and 0.1 mM DTT. Cell debris was pelleted by centrifugation at 20,000 x g for 15 min and the soluble lysate was incubated with anti-Flag magnetic beads for 2 h at 4 °C. After washing, Flag-tagged and co-precipitated, proteins were eluted by addition of 250 μM Flag peptide for 30 min at 12 °C. Co-precipitated proteins were detected by mass spectrometry or by western blotting using antibodies against NOP14 (Bethyl; A303-869A), NOC4L (Novus Biologicals; NBP1-92191), UTP14A (Proteintech; 11474-1-AP) and WDR3 (Bethyl; A301-553A). Alternatively, 80 μg His-ZZ-tagged EMG1 was immobilized on pre-equilibrated IgG sepharose in 50 mM Tris pH 7.4, 150 mM NaCl, 2.5 MgCl2 and 2.5 mM beta-mercaptoethanol. 0.5 ml HeLa extract in 10 mM HEPES/KOH pH 7.6, 10 mM KOAc, 0.5 mM MgOAc, 5 mM DTT and protease inhibitor cocktail (cOmplete; Roche) was pre-treated with 100 mM NaCl and centrifuged at 21,000 x g at 4 °C to precipitate insoluble components. The cleared lysate was either pre-treated with 2 μM RNase A at 4 °C for 2 h or was incubated directly with immobilised His-ZZ-tagged EMG1 for 3 h at room temperature. The beads were then washed and elution was performed by treatment with 2 μM TEV protease for 4 h at room temperature in 50 mM Tris-HCl pH 7.8, 150 mM NaCl, 1.5 mM MgCl2, 0.1% NP40 and 5 mM β-mercaptoethanol. Samples were then analysed by SDS-PAGE and western blotting using antibodies against NOP14, NOC4L, UTP14A and MPP10 (51).
Human cell culture and generation of stable cell lines
HEK293 Flp-In T-Rex (Life Technologies) and HeLa CCL2 cells were grown at 37 °C and 5% CO2 in DMEM medium (Invitrogen) supplemented with 10% FCS, 10 μg/ml penicillin, 10 μg/ml streptomycin and 2 mM glutamine. To generate doxycycline-inducible stable cell lines the EMG1 coding sequence (NM_006331.7) was first cloned into the pcDNA5 vector with no tag or with an N-terminal 2xFlag-PreScission protease cleavage site-His6 tag. For expression of siRNA-insensitive EMG1 five silent mutations were introduced into the siRNA target site by site-directed mutagenesis and to enable expression EMG1 with of the BCS mutation a single point mutation was introduced to encode glycine in position 86. HEK293 Flp-In T-Rex cells (Life Technologies) were transfected with the plasmids according to manufacturer’s instructions. Expression of the transgene was induced using up to 1 ng/ml doxycycline for 24 h.
RNA interference, RNA extraction and qRT-PCR
Cells were transfected with siRNAs (see Supplementary Material, Table S1) using Lipofectamine (RNAiMAX; Life Technologies) according to manufacturer’s instructions. Total RNA was isolated from cells 72 h after siRNA transfection using TRIreagent (Sigma) and cDNA was synthesised using SuperScript III reverse transcriptase (Life Technologies) according to the manufacturer’s instructions. qRT-PCR and relative quantification using levels of the GAPDH mRNA for reference were performed as described (52) with the primers EMG1_CDS_fw 5’-CCACCAGAGTTTGCTGATGC-3’ and EMG1_CDS_rev 5’-ATTCGGGTCTGGGGATT CAC-3’ for the detection of both the endogenous and the tet-induced EMG1 mRNAs, EMG1_3UTR_fw 5’-GCCAAGGCTGTACATTTGCT-3’ and EMG1_3UTR_rev 5’-TAGGAACCCCAAAACAACCAGG-3’ for the amplification of only the endogenous EMG1 mRNA and GAPDH_fw 5’-GAGGGTCTCTCTCTTCCTC-3’ and GAPDH_rev 5’-CAACGTGTCAGTGGTGGAC-3’.
Northern blotting
Pre-rRNAs were detected with Northern blotting as described previously (53) using specific probes (5’ ITS1 5’-CCTCGCCCTCCGGGCTCCGTTAATGATC-3’; actin mRNA 5’-AGGGATAGCACAGCCTGGATAGCAAC-3’).
Pulse labelling
Metabolic labelling experiments using 32P-orthophosphate were performed as previously described (54). In brief, siRNA-treated cells were grown in phosphate-free Dulbecco’s modified eagles medium (DMEM) for 1 h and then in DMEM supplemented with 15 μCi/ml 32P-orthophosphate for a further 1 h (pulse). After growth for an additional 3 h (chase) in unlabelled DMEM, total RNA was extracted using TRIreagent (Sigma) and analysed by agarose-glyoxal gel electrophoresis. RNA was transferred to a nylon membrane and newly synthesised RNAs were detected using a phosphorimager.
Immunofluorescence
Immunofluorescence was basically performed as previously described (55,56). Cells were grown on coverslips and fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min. Cells were then permeabilised with 0.1% triton-X-100 in PBS for 15 min and blocked with 10% foetal calf serum (FCS) and 0.1% triton-X-100 in PBS for 1 h at room temperature. Cells were then incubated with primary antibodies against EMG1 (this study), NOP14, UTP14A and NSUN5 (Santa Cruz Biotechnology; sc-376147) in 10% FCS and PBS overnight at 4 °C. Cells were washed and incubated with secondary antibodies (Alexa Fluor 488-conjugated donkey anti-guinea pig, Alexa Fluor 594-conjugated donkey anti-mouse, Alexa Fluor 594-conjugated donkey anti-rabbit or Alexa Fluor 647-conjugated donkey anti-rabbit) for 2 h at room temperature. Coverslips were mounted onto slides using a mounting medium with DAPI (Vectashield; Vector Laboratories) and fluorescence was analysed using an inverted confocal microscope (ConfoCor 2; Carl Zeiss).
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank Uwe Plessmann and Henning Urlaub for mass spectrometry, Claudia Höbartner for the RNA oligonucleotides used for in vitro methylation assays and Nicholas Watkins for the MPP10 antibody. This work was supported by the Deutsche Forschungsgemeinschaft (SFB1190 to M.T.B. and D.G.); the Max Planck Society (to D.G.); the Faculty of Medicine, Georg-August-University Göttingen (M.T.B. and ‘Startförderung’ to S.H.); and the Alexander von Humboldt foundation (to K.E.S. and M.T.B).
Conflict of Interest statement. None declared.
Funding
This work was supported by the Deutsche Forschungsgemeins-chaft (SFB1190 to M.T.B. and D.G.); the Max Planck Society (to D.G.); the Faculty of Medicine, Georg-August-University Göttingen (to M.T.B. and “Startförderung” to S.H.) and the Alexander von Humboldt Foundation (to K.E.S. and M.T.B.). Funding to pay the Open Access publication charges for this article was provided by the Deutsche Forschungsgemeinschaft (SFB1190).
References
- 1.Warner J.R. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci., 24, 437–440. [DOI] [PubMed] [Google Scholar]
- 2.Donati G., Montanaro L., Derenzini M. (2012) Ribosome biogenesis and control of cell proliferation: p53 is not alone. Cancer Res., 72, 1602–1607. [DOI] [PubMed] [Google Scholar]
- 3.Woolford J.L., Jr., Baserga S.J. (2013) Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics, 195, 643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watkins N.J., Bohnsack M.T. (2012) The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA, 3, 397–414. [DOI] [PubMed] [Google Scholar]
- 5.Henras A.K., Plisson-Chastang C., O'Donohue M.F., Chakraborty A., Gleizes P.E. (2015) An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA, 6, 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma S., Lafontaine D.L. (2015) ′View From A Bridge′: A New Perspective on Eukaryotic rRNA Base Modification. Trends Biochem. Sci., 40, 560–575. [DOI] [PubMed] [Google Scholar]
- 7.Decatur W.A., Fournier M.J. (2002) rRNA modifications and ribosome function. Trends Biochem. Sci., 27, 344–351. [DOI] [PubMed] [Google Scholar]
- 8.Wild T., Horvath P., Wyler E., Widmann B., Badertscher L., Zemp I., Kozak K., Csucs G., Lund E., Kutay U. (2010) A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol., 8, e1000522.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin R., Straub A.U., Doebele C., Bohnsack M.T. (2013) DExD/H-box RNA helicases in ribosome biogenesis. RNA Biol., 10, 4–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tafforeau L., Zorbas C., Langhendries J.L., Mullineux S.T., Stamatopoulou V., Mullier R., Wacheul L., Lafontaine D.L. (2013) The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol. Cell, 51, 539–551. [DOI] [PubMed] [Google Scholar]
- 11.Badertscher L., Wild T., Montellese C., Alexander L.T., Bammert L., Sarazova M., Stebler M., Csucs G., Mayer T.U., Zamboni N., et al. (2015) Genome-wide RNAi Screening Identifies Protein Modules Required for 40S Subunit Synthesis in Human Cells. Cell Rep., 13, 2879–2891. [DOI] [PubMed] [Google Scholar]
- 12.Narla A., Ebert B.L. (2010) Ribosomopathies: human disorders of ribosome dysfunction. Blood, 115, 3196–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armistead J., Triggs-Raine B. (2014) Diverse diseases from a ubiquitous process: the ribosomopathy paradox. FEBS Lett., 588, 1491–1500. [DOI] [PubMed] [Google Scholar]
- 14.Sondalle S.B., Baserga S.J. (2014) Human diseases of the SSU processome. Biochim. Biophys. Acta, 1842, 758–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freed E.F., Bleichert F., Dutca L.M., Baserga S.J. (2010) When ribosomes go bad: diseases of ribosome biogenesis. Mol. Biosyst., 6, 481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fumagalli S., Thomas G. (2011) The role of p53 in ribosomopathies. Semin. Hematol., 48, 97–105. [DOI] [PubMed] [Google Scholar]
- 17.Ellis S.R., Gleizes P.E. (2011) Diamond Blackfan anemia: ribosomal proteins going rogue. Semin. Hematol., 48, 89–96. [DOI] [PubMed] [Google Scholar]
- 18.Ellis S.R. (2014) Nucleolar stress in Diamond Blackfan anemia pathophysiology. Biochim. Biophys. Acta, 1842, 765–768. [DOI] [PubMed] [Google Scholar]
- 19.Dixon J., Trainor P., Dixon M.J. (2007) Treacher Collins syndrome. Orthod Craniofac Res, 10, 88–95. [DOI] [PubMed] [Google Scholar]
- 20.Hannan K.M., Sanij E., Rothblum L.I., Hannan R.D., Pearson R.B. (2013) Dysregulation of RNA polymerase I transcription during disease. Biochim. Biophys. Acta, 1829, 342–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freed E.F., Prieto J.L., McCann K.L., McStay B., Baserga S.J. (2012) NOL11, implicated in the pathogenesis of North American Indian childhood cirrhosis, is required for pre-rRNA transcription and processing. PLoS Genet., 8, e1002892.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffin J.N., Sondalle S.B., Del Viso F., Baserga S.J., Khokha M.K. (2015) The ribosome biogenesis factor Nol11 is required for optimal rDNA transcription and craniofacial development in Xenopus. PLoS Genet., 11, e1005018.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finch A.J., Hilcenko C., Basse N., Drynan L.F., Goyenechea B., Menne T.F., González Fernández A., Simpson P., D'Santos C.S., Arends M.J., et al. (2011) Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev., 25, 917–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weis F., Giudice E., Churcher M., Jin L., Hilcenko C., Wong C.C., Traynor D., Kay R.R., Warren A.J. (2015) Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat. Struct. Mol. Biol., 22, 914–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowen P., Conradi G.J. (1976) Syndrome of skeletal and genitourinary anomalies with unusual facies and failure to thrive in Hutterite sibs. Birth Defects Orig. Artic Ser., 12, 101–108. [PubMed] [Google Scholar]
- 26.Hunter A.G., Woerner S.J., Montalvo-Hicks L.D., Fowlow S.B., Haslam R.H., Metcalf P.J., Lowry R.B. (1979) The Bowen-Conradi syndrome – a highly lethal autosomal recessive syndrome of microcephaly, micrognathia, low birth weight, and joint deformities. Am. J. Med. Genet., 3, 269–279. [DOI] [PubMed] [Google Scholar]
- 27.Lowry R.B., Innes A.M., Bernier F.P., McLeod D.R., Greenberg C.R., Chudley A.E., Chodirker B., Marles S.L., Crumley M.J., Loredo-Osti J.C., et al. (2003) Bowen–Conradi syndrome: a clinical and genetic study. Am. J. Med. Genet., 120A, 423–428. [DOI] [PubMed] [Google Scholar]
- 28.Armistead J., Khatkar S., Meyer B., Mark B.L., Patel N., Coghlan G., Lamont R.E., Liu S., Wiechert J., Cattini P.A., et al. (2009) Mutation of a gene essential for ribosome biogenesis, EMG1, causes Bowen–Conradi syndrome. Am. J. Hum. Genet., 84, 728–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebersberger I., Simm S., Leisegang M.S., Schmitzberger P., Mirus O., von Haeseler A., Bohnsack M.T., Schleiff E. (2014) The evolution of the ribosome biogenesis pathway from a yeast perspective. Nucleic Acids Res., 42, 1509–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu P.C., Thiele D.J. (2001) Novel stress-responsive genes EMG1 and NOP14 encode conserved, interacting proteins required for 40S ribosome biogenesis. Mol. Biol. Cell, 12, 3644–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eschrich D., Buchhaupt M., Kotter P., Entian K.D. (2002) Nep1p (Emg1p), a novel protein conserved in eukaryotes and archaea, is involved in ribosome biogenesis. Curr. Genet., 40, 326–338. [DOI] [PubMed] [Google Scholar]
- 32.Meyer B., Wurm J.P., Kötter P., Leisegang M.L., Schilling V., Buchhaupt M., Held M., Bahr U., Karas M., Heckel A., et al. (2011) The Bowen-Conradi syndrome protein Nep1 (Emg1) has a dual role in eukaryotic ribosome biogenesis, as an essential assembly factor and in the methylation of 1191 in yeast 18S rRNA. Nucleic Acids Res., 39, 1526–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kornprobst M., Turk M., Kellner N., Cheng J., Flemming D., Koš-Braun I., Koš M., Thoms M., Berninghausen O., Beckmann R., Hurt E. (2016) Architecture of the 90S Pre-ribosome: A Structural View on the Birth of the Eukaryotic Ribosome. Cell, 166, 380–393. [DOI] [PubMed] [Google Scholar]
- 34.Meyer B., Wurm J.P., Sharma S., Immer C., Pogoryelov D., Kötter P., Lafontaine D.L., Wöhnert J., Entian K.D. (2016) Ribosome biogenesis factor Tsr3 is the aminocarboxypropyl transferase responsible for 18S rRNA hypermodification in yeast and humans. Nucleic Acids Res., 44, 4304–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leulliot N., Bohnsack M.T., Graille M., Tollervey D., Van Tilbeurgh H. (2008) The yeast ribosome synthesis factor Emg1 is a novel member of the superfamily of alpha/beta knot fold methyltransferases. Nucleic Acids Res., 36, 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haag S., Kretschmer J., Bohnsack M.T. (2015) WBSCR22/Merm1 is required for late nuclear pre-ribosomal RNA processing and mediates N7-methylation of G1639 in human 18S rRNA. RNA, 21, 180–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor A.B., Meyer B., Leal B.Z., Kotter P., Schirf V., Demeler B., Hart P.J., Entian K.D., Wöhnert J. (2008) The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site. Nucleic Acids Res., 36, 1542–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas S.R., Keller C.A., Szyk A., Cannon J.R., Laronde-Leblanc N.A. (2010) Structural insight into the functional mechanism of Nep1/Emg1 N1-specific pseudouridine methyltransferase in ribosome biogenesis. Nucleic Acids Res., 39, 2445–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kühn H., Hierlmeier T., Merl J., Jakob S., Aguissa-Touré A.H., Milkereit P., Tschochner H. (2009) The Noc-domain containing C-terminus of Noc4p mediates both formation of the Noc4p-Nop14p submodule and its incorporation into the SSU processome. PLoS One, 4, e8370.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jäkel S., Mingot J.M., Schwarzmaier P., Hartmann E., Görlich D. (2002) Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. Embo J., 21, 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jäkel S., Albig W., Kutay U., Bischoff F.R., Schwamborn K., Doenecke D., Görlich D. (1999) The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. Embo J., 18, 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bäuerle M., Doenecke D., Albig W. (2002) The requirement of H1 histones for a heterodimeric nuclear import receptor. J. Biol. Chem., 277, 32480–32489. [DOI] [PubMed] [Google Scholar]
- 43.Armistead J., Hemming R., Patel N., Triggs-Raine B. (2014) Mutation of EMG1 causing Bowen-Conradi syndrome results in reduced cell proliferation rates concomitant with G2/M arrest and 18S rRNA processing delay. BBA Clin., 1, 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Armistead J., Patel N., Wu X., Hemming R., Chowdhury B., Basra G.S., Del Bigio M.R., Ding H., Triggs-Raine B. (2015) Growth arrest in the ribosomopathy, Bowen-Conradi syndrome, is due to dramatically reduced cell proliferation and a defect in mitotic progression. Biochim. Biophys. Acta, 1852, 1029–1037. [DOI] [PubMed] [Google Scholar]
- 45.Wurm J.P., Meyer B., Bahr U., Held M., Frolow O., Kötter P., Engels J.W., Heckel A., Karas M., Entian K.D., Wöhnert J. (2010) The ribosome assembly factor Nep1 responsible for Bowen-Conradi dyndrome is a pseudouridine-N1-specific methyltransferase. Nucleic Acids Res., 38, 2387–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kressler D., Bange G., Ogawa Y., Stjepanovic G., Bradatsch B., Pratte D., Amlacher S., Strauß D., Yoneda Y., Katahira J., et al. (2012) Synchronizing nuclear import of ribosomal proteins with ribosome assembly. Science, 338, 666–671. [DOI] [PubMed] [Google Scholar]
- 47.Stelter P., Huber F.M., Kunze R., Flemming D., Hoelz A., Hurt E. (2015) Coordinated Ribosomal L4 Protein Assembly into the Pre-Ribosome Is Regulated by Its Eukaryote-Specific Extension. Mol. Cell, 58, 854–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pillet B., García-Gómez J.J., Pausch P., Falquet L., Bange G., de la Cruz J., Kressler D. (2015) The dedicated chaperone Acl4 escorts ribosomal protein Rpl4 to its nuclear Pre-60S assembly site. PLoS Genet., 11, e1005565.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jäkel S., Görlich D. (1998) Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. Embo J., 17, 4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frey S., Görlich D. (2014) Purification of protein complexes of defined subunit stoichiometry using a set of orthogonal, tag-cleaving proteases. J. Chromatogr. A, 1337, 106–115. [DOI] [PubMed] [Google Scholar]
- 51.Turner A.J., Knox A.A., Prieto J.L., McStay B., Watkins N.J. (2009) A novel small-subunit processome assembly intermediate that contains the U3 snoRNP, nucleolin, RRP5, and DBP4. Mol. Cell Biol., 29, 3007–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sloan K.E., Bohnsack M.T., Schneider C., Watkins N.J. (2014) The roles of SSU processome components and surveillance factors in the initial processing of human ribosomal RNA. RNA, 20, 540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sloan K.E., Leisegang M.S., Döbele C., Ramirez A.S., Simm S., Safferthal C., Kretschmer J., Schorge T., Markoutsa S., Haag S., et al. (2015) The association of late-acting snoRNPs with human pre-ribosomal complexes requires the RNA helicase DDX21. Nucleic Acids Res., 43, 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sloan K.E., Mattijssen S., Lebaron S., Tollervey D., Pruijn G.J.M., Watkins N.J. (2013) Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J. Cell Biol., 200, 577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haag S., Warda A.S., Kretschmer J., Günnigmann M.A., Höbartner C., Bohnsack M.T. (2015) NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA, 21, 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haag S., Sloan K.E., Ranjan N., Warda A.S., Kretschmer J., Blessing C., Hübner B., Seikowski J., Dennerlein S., Rehling P., et al. (2016) NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. Embo J., 35, 2104–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.