Figure 2.
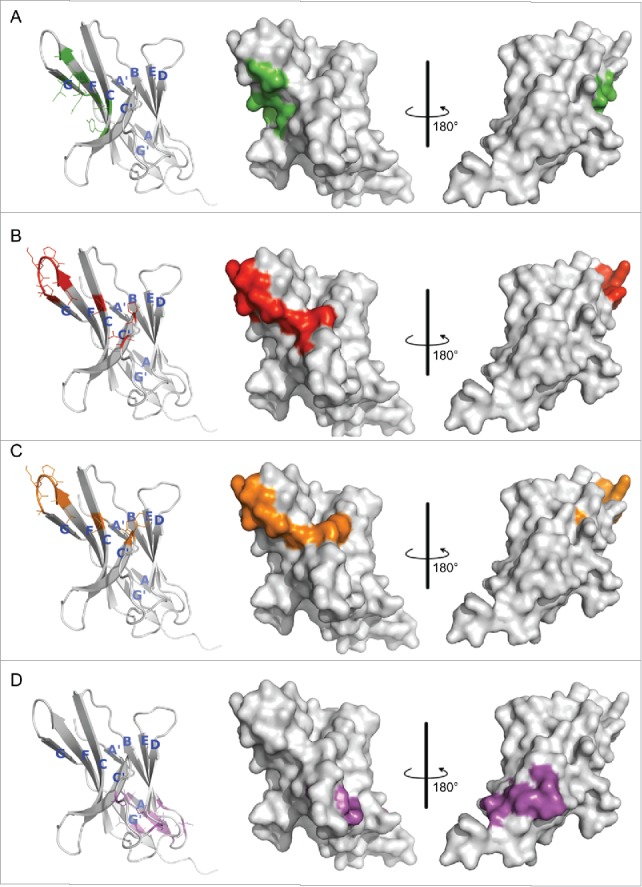
Hot spot residues mapped on hPD-1 structure. (A) hPD-L1 binding site. Data were obtained from the literature.18 (B-D) Binding sites of antibody R11, R9 and the control antibody R10, respectively. Data were from Table 1. Colors in the pictures are included to help distinguish differences between epitopes. The structure of hPD-1 was taken from the crystal structure (PDB code 4ZQK), whose missing loops (Asp85-Asp92) were remodeled by adopting initial conformations from the NMR structure (PDB code 2M2D). The back β-strands are A′, B, E, D, A and G′. The front sheets are labeled in G, F, C and C′. The C″ strand observed in mPD-1 lost its secondary structure in hPD-1.18, 20
