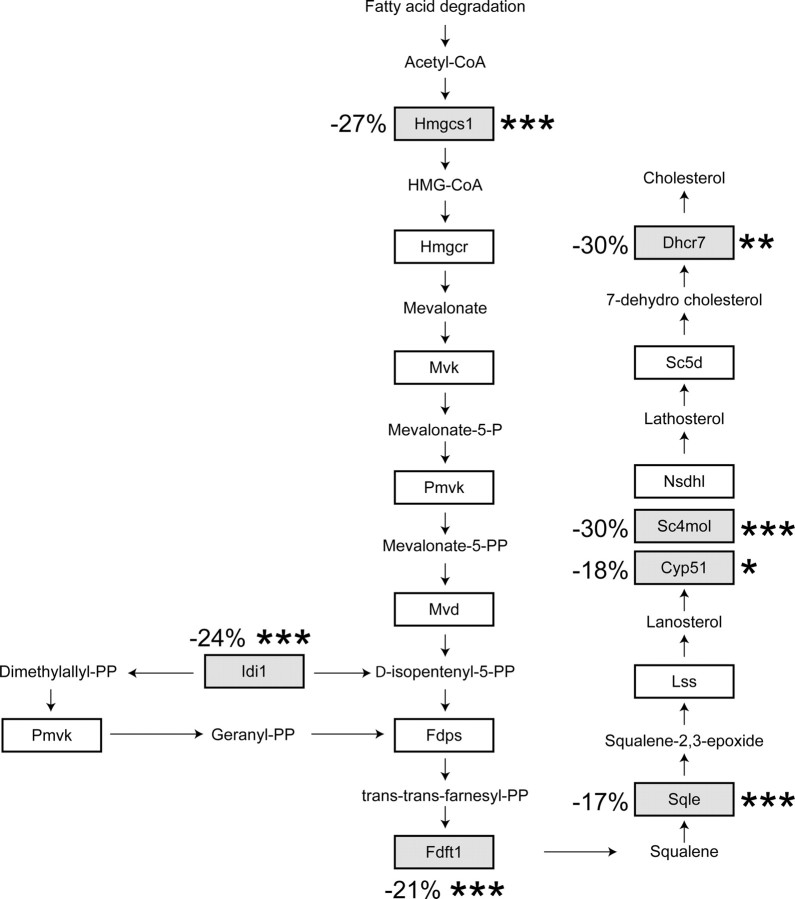
Fig. 2.
HDL effect on cholesterol synthesis genes. The figure shows enzymes involved in cholesterol synthesis that are regulated by SREBPs. Those the mRNAs of which were found to be negatively modulated by HDLs in the transcriptomic analysis are in gray boxes. The numbers correspond to the decrease in mRNAs levels induced by HDLs in starved cells compared with the values obtained from control starved cells. Asterisks indicate when the decrease is statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Hmgcs1, 3-Hydroxy-3-methyl-glutaryl (HMG)-CoA synthase 1; Hmgcr, HMG-CoA reductase; Mvk, mevalonate kinase; Pmvk, phospho-mevalonate kinase; Mvd, mevalonate pyrophosphate decarboxylate; Idi1, isopentenyl-diphosphate Δ-isomerase 1; Fdps, farnesyl diphosphate synthase; Fdft1, farnesyl diphosphate farnesyl transferase 1; Sqle, squalene epoxidase; Lss, lanosterol synthase; CYP51, lanosterol 14α-demethylase; Sc4 mol, sterol-C4-methyl oxidase-like; Nsdhl, nicotinamide adenine dinucleotide (phosphate)-dependent steroid dehydrogenase-like; Sc5d, sterol C5-desaturase; Dhcr7, 7-dehydrocholesterol reductase.