Fig. 3.
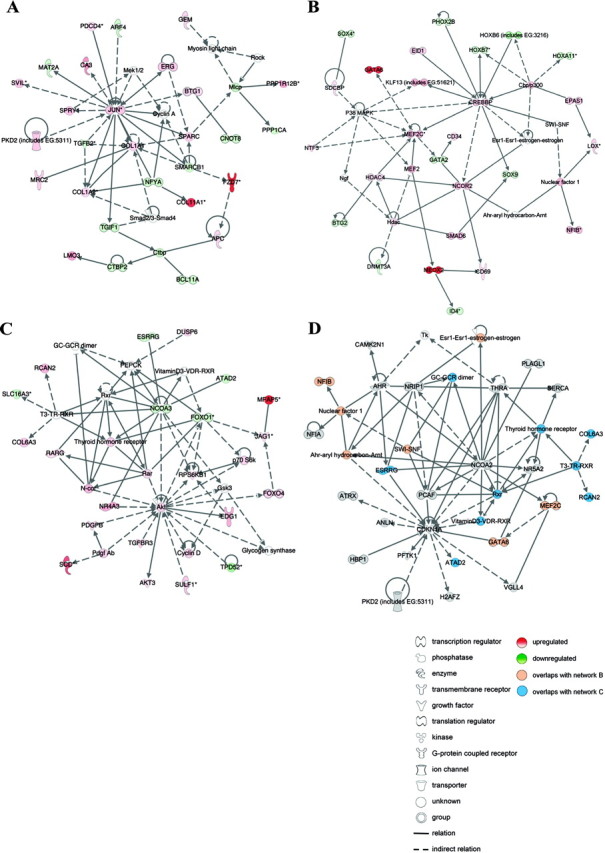
Functional analysis of differentially expressed miRNA targets in endometriosis. A–C, Networks identified by IPA of differentially expressed mRNAs that are also predicted targets of the 22 differentially expressed miRNAs in paired peritoneal ectopic vs. eutopic endometrial tissues from women with endometriosis. A, c-Jun is central in a network comprising extracellular matrix proteins that are confirmed targets of miRNAs differentially expressed in endometriosis. This network overlapped considerably to analysis of all predicted targets of the 22 miRNAs (Fig. 2C), suggesting a role for miRNAs in regulating cell migration during endometrial lesion development. B, MEF2C, histone deacetylase 4 (HDAC4), and DNA (cytosine-5-) methyltransferase 3α (DNMT3A) are validated miRNA targets represented in a network converging on CREBBP, proposing a role for miRNAs in the regulation of myogenesis, angiogenesis, and epigenetic modulation of gene expression in endometrial tissue remodeling. C, The IPA network converging on AKT indicated a role for miRNAs in regulating retinoic acid signaling, the cell cycle, and cellular growth and proliferation. D, IPA network from functional analysis of a second microarray data set of differentially expressed miRNA targets in paired ovarian ectopic vs. eutopic endometrial tissue from women with endometriosis. The network exemplifies the substantial overlap between the ovarian and the peritoneal comparisons, illustrated in orange and blue nodes. Red and green nodes depict up- and down-regulated miRNA targeted transcripts in ectopic vs. eutopic endometrial tissue, respectively. Lines represent the biological relationship between two nodes. The gene names in these networks may be identified on the HUGO Gene Nomenclature Committee home page (http://www.genenames.org).
