Fig. 2.
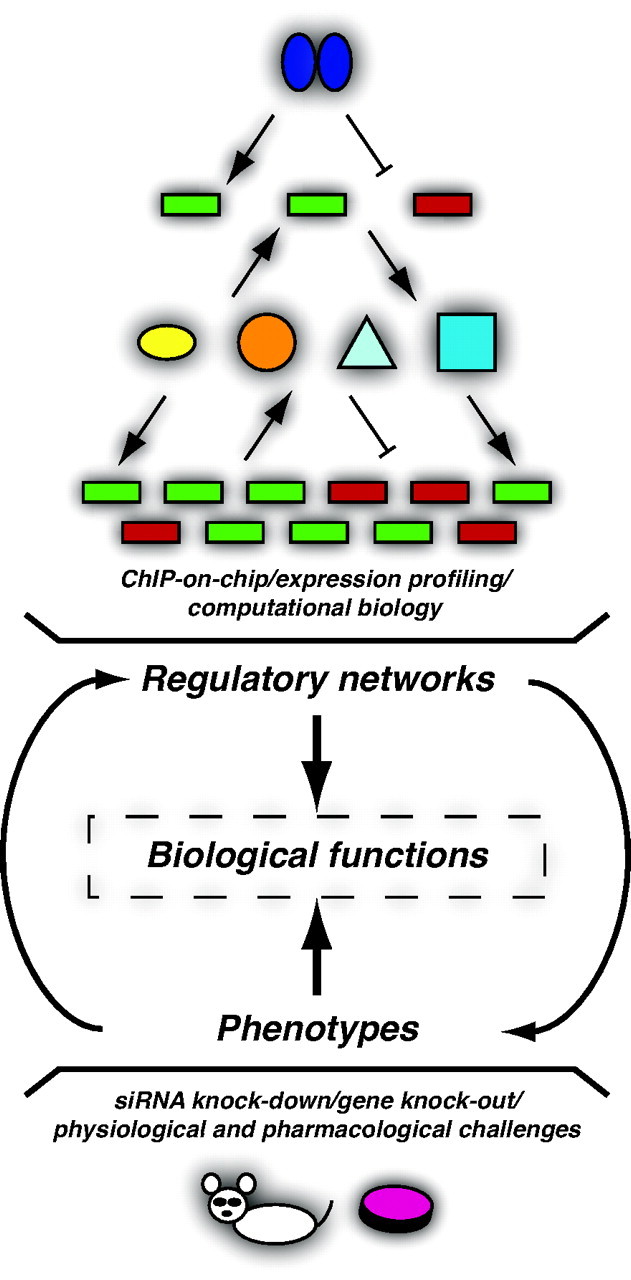
Regulatory Networks, Phenotypic Analyses, and Biological Functions of Nuclear Receptors
Nuclear receptors control the expression of a limited set of primary target genes. Some of these genes encode transcription factors, coregulator proteins, micro-RNAs, and posttranslational modifiers of proteins that will both influence the original primary response and lead to a broad secondary response. ChIP-based, expression profiling, and computational biology approaches are considered a useful set of tools with which to build regulatory networks (top). In parallel, phenotypic analyses using small interfering RNA knockdown, gene knock-out and overexpression, as well as physiological and pharmacological challenges in cell-based and animal models are used to study the overall functions of a particular nuclear receptor (bottom). When integrated, the two complementary approaches can lead to the identification of nuclear receptor-dependent biological functions for which we have a clear understanding of the molecular mechanisms underlying the observed phenotypes (middle). A nuclear receptor is represented by two blue ovals; genes are represented by green and red boxes for up- and down-regulated genes, respectively; gene products are represented by diverse shapes and colors. Arrows indicate activation; blunt arrows represent repression. siRNA, small interfering RNA.
