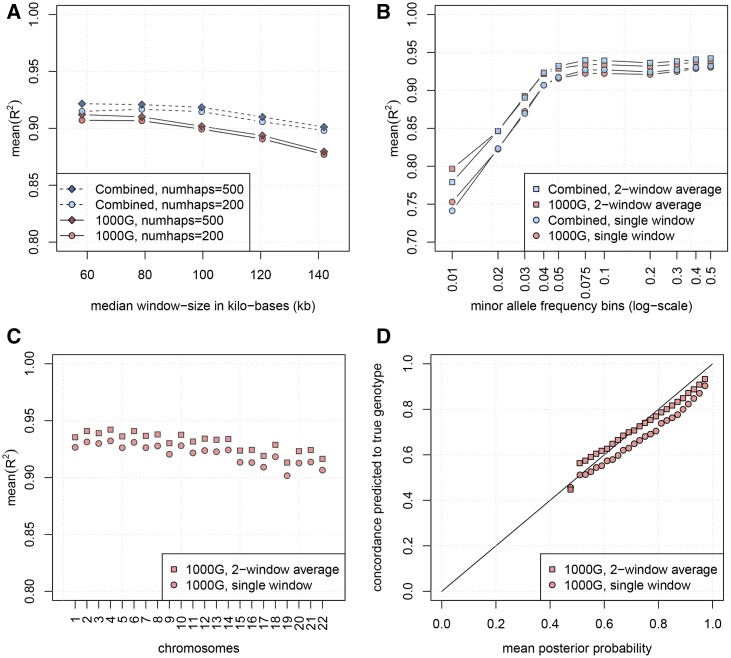
Figure 1.
Mean- and calibration for imputation based on GeneImp. (A) Mean- as a function of window-size. Results are from chromosome 22. A smaller window and the Combined panel lead to higher mean- while more filtered haplotypes lead to very small gains. (B) Mean- as a function of MAF. Results are from the whole genome using filtered haplotypes. Single window-split corresponds to median window-size of 58.2 kb, average of two window-splits is taken over results with median window-sizes of 58.2 and 78.9 kb. Mean- increases as a function of the MAF, leveling-off around Averaging posterior probabilities from two window-splits leads to higher mean- especially for rarer SNPs. (C) Mean- in different chromosomes. Results are based on filtered haplotypes. Single window-split corresponds to median window-size of 58.2 kb, average of two window-splits is taken over results with median window-sizes of 58.2 and 78.9 kb. Imputation is marginally worse in shorter chromosomes. (D) Calibration of posterior probabilities from a single window-split corresponding to median window-size of 58.2 kb, and an average of two window-splits taken over results from median window-sizes of 58.2 and 78.9 kb. To evaluate calibration we split imputed genotypes into bins according to their posterior probability distribution. We plot the mean posterior probability in each bin (x-axis) against the percentage of correctly predicted genotypes in each bin (y-axis). Averaging across window-splits leads to well calibrated posterior probabilities (most points lie close to the diagonal), while imputation probabilities based on a single window-split are over-confident (points lie below the diagonal).