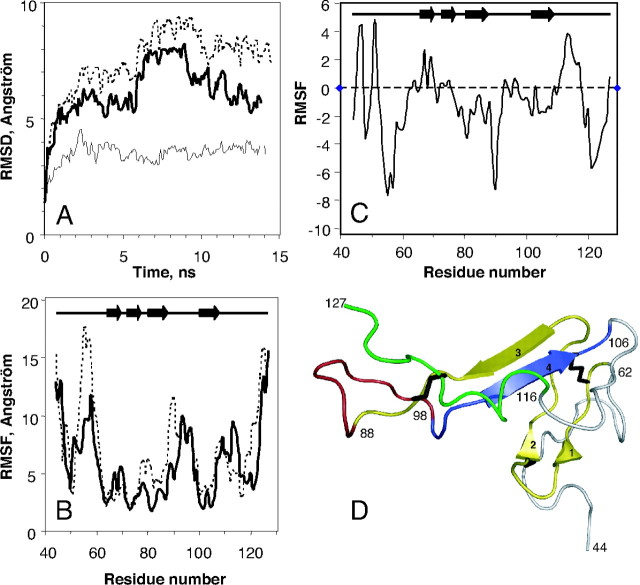
Fig. 5.
Molecular Dynamic Simulations of hVPAC1 N-ted in the Presence or the Absence of VIP
A, Conformational drift from the initial structure during the simulations, measured as backbone RMSD for the hVPAC1 N-ted in the presence (dashed line) and in the absence (bolded line) of VIP. The same data are also shown for VIP itself (thin line) docked in the receptor N-ted. Plots were drawn using a 0.1-nsec sampling. B, Conformational flexibility of hVPAC1 N-ted measured as RMSF of the backbone atoms during the 14-nsec molecular dynamic simulation as a function of residue number. Data are shown for hVPAC1 N-ted in the presence (bolded line), and the absence (dashed line) of VIP. C, Conformational flexibility of hVPAC1 N-ted due to the presence of VIP measured as ΔRMSF, which represents difference between the RMSFs of hVPAC1 N-ted in the presence and in the absence of VIP. D, Several segments of the receptor N-ted were colored to more easily follow the text in Results and Discussion. Yellow, 63–88 segment; red, 89–98 segment; blue, 99–106 segment; green, 116–127 segment. The three disulfide bridges (C50-C72, C63-C105, and C86-C122) were represented in black. Panel D was produced using Pymol software (http://www.pymol.org). ns, Nanosecond.