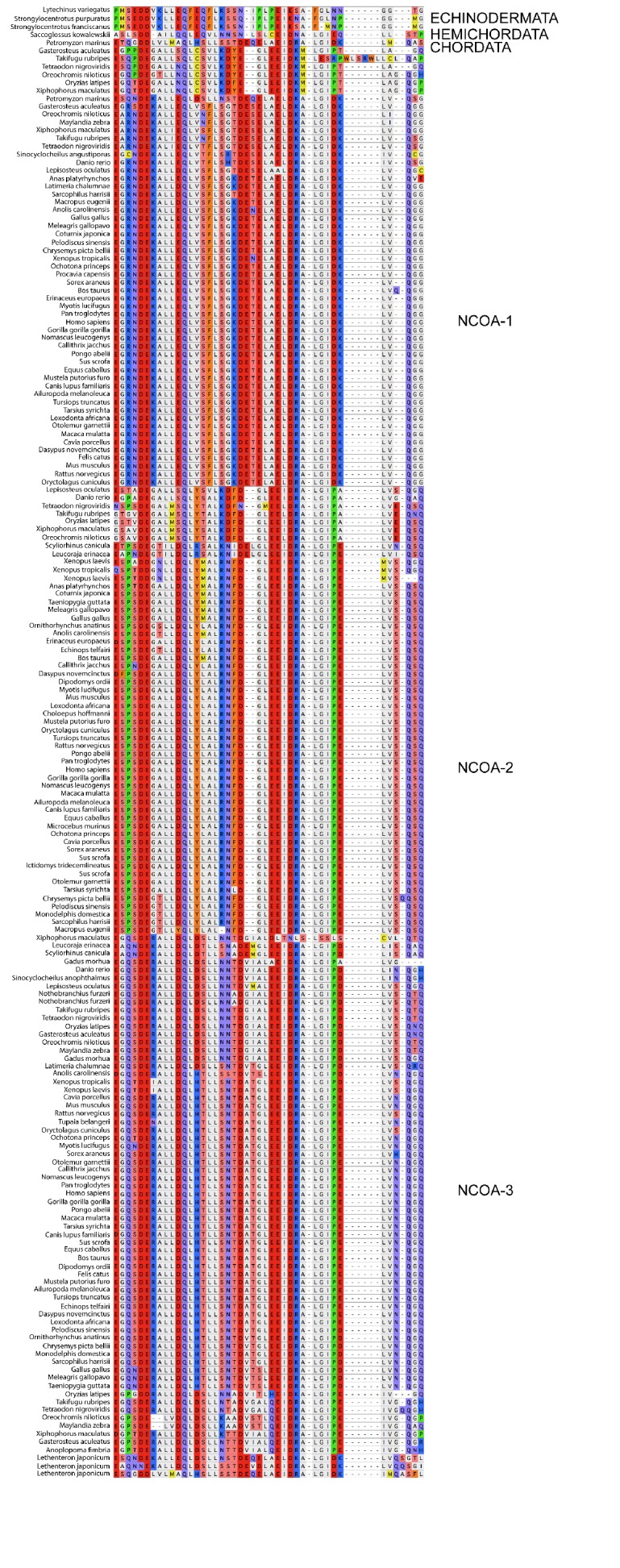
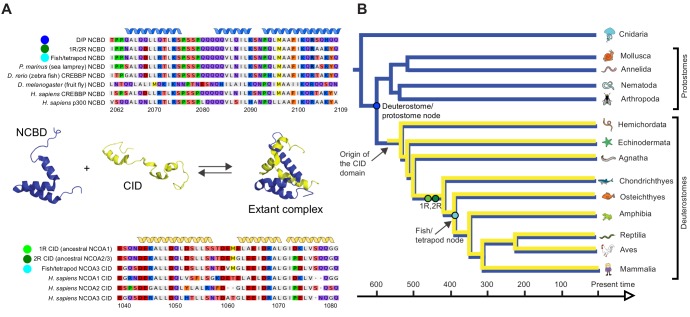
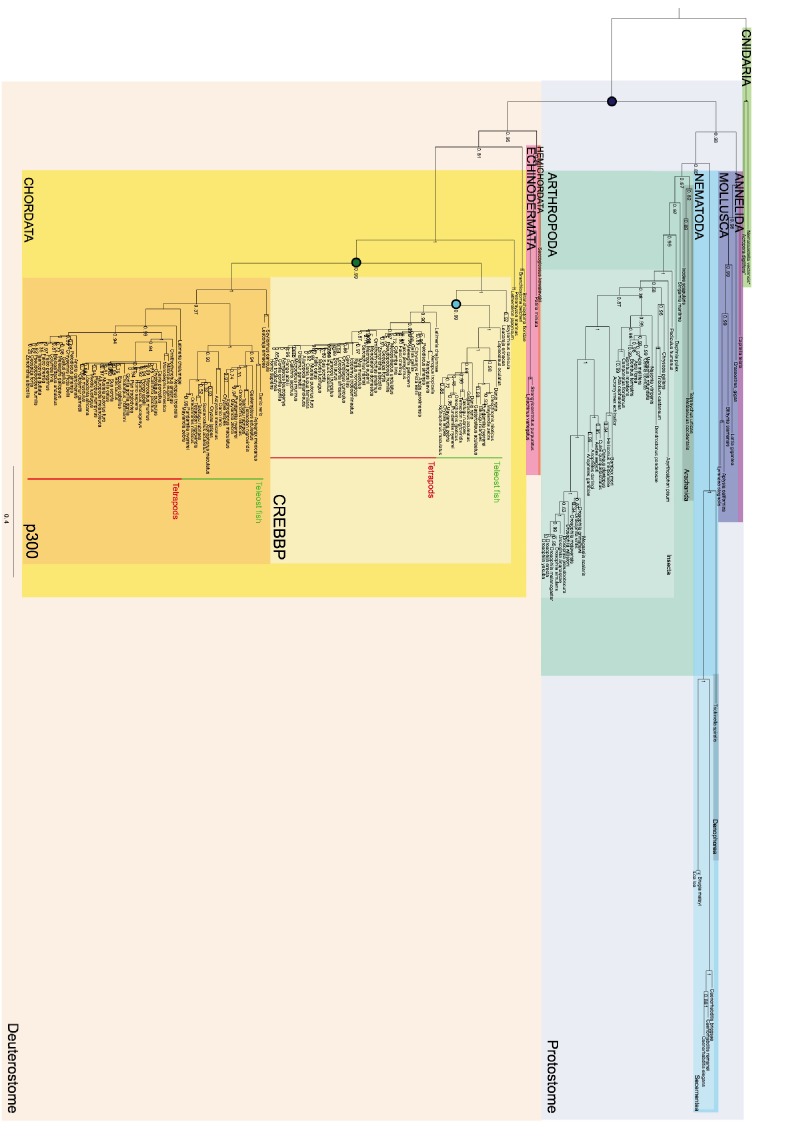
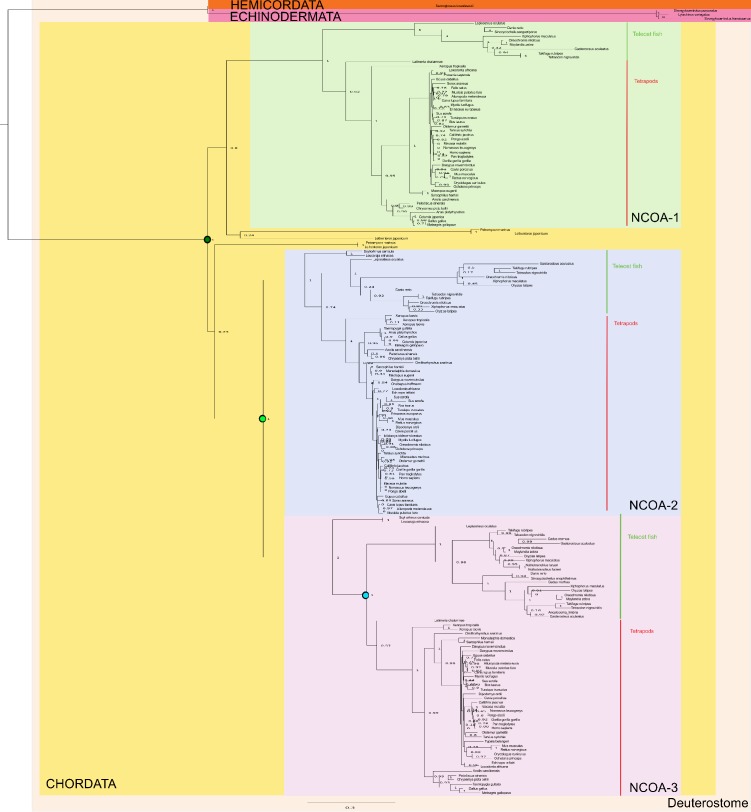
Figure 2. Reconstruction of the evolution of the interacting NCBD and CID domains.
(A) Sequence alignments of extant and reconstructed ancient NCBD (top) and CID domains (bottom). The positions of helices are according to the NMR structure of the complex between extant CREBBP NCBD (blue) and NCOA3 CID (yellow). Free NCBD (protein data base code 2KKJ) and the CID/NCBD complex (1KBH) are NMR structures, whereas the picture of free CID is a hypothetical modified structure made from the NCOA1 CID/NCBD complex (2C52). The first residue in the NCBD alignment is referred to as position 2062 in the text and the first residue in the CID alignment as 1040. The color coding of the sequences reflects similarities in chemical properties of the amino acid side chains and is a guide for the eye to see patterns of conservation. (B) Schematic tree of life with selected animal groups depicting the evolution of the NCBD domain (blue) in both protostomes and deuterostomes and the CID domain (yellow) in the deuterostome lineage only. See Figure 2—figure supplements 1–4 for detailed alignments and trees.
DOI: http://dx.doi.org/10.7554/eLife.16059.004
Figure 2—figure supplement 1. Sequence alignment of NCBD domains of CREBBP/p300 used in the phylogenetic reconstruction.
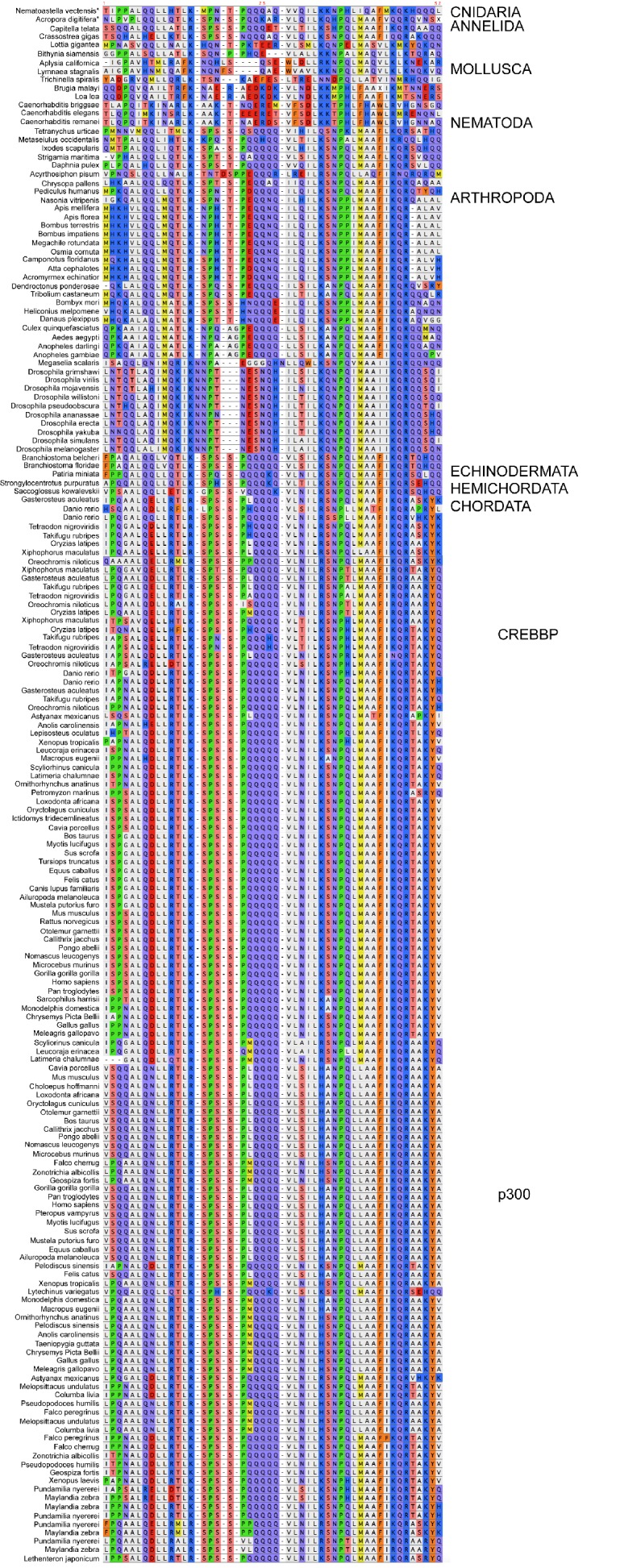
Figure 2—figure supplement 2. Sequence alignment of the CID domains of NCOA1-3 used in the phylogenetic reconstruction.