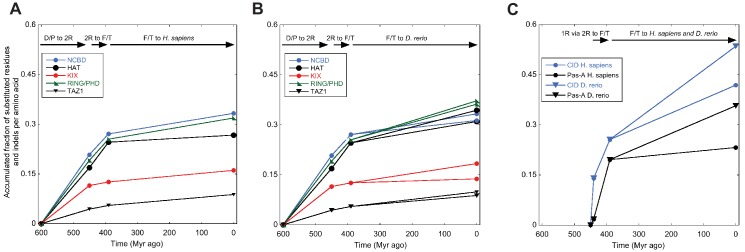
Figure 3. Amino acid substitutions in different domains in CREBBP/p300 and NCOA as a function of time.
The predicted ancient sequences for distinct domains in CREBBP/p300 (A and B) and NCOA (C) were used to calculate the number of substitutions and indels between each evolutionary node (Deuterostome/protostome, D/P; 1R; 2R; Fish/tetrapod, F/T; and present day) in a particular lineage (human and zebrafish CREBBP and human and zebrafish NCOA3, respectively). The alignment and trees used to resurrect HAT, KIX, RING/PHD and TAZ1 were the ones optimized for NCBD. Similarly, the alignment and trees used to resurrect Pas-A were the ones optimized for the CID domain. The number of substitutions plus indels were normalized against the number of amino acid residues in each domain and the accumulated fraction of sequence changes plotted versus historical time. Both 1R and 2R occurred around 450 million Myr ago and the distance between them in panel C (10 Myr) is arbitrary.