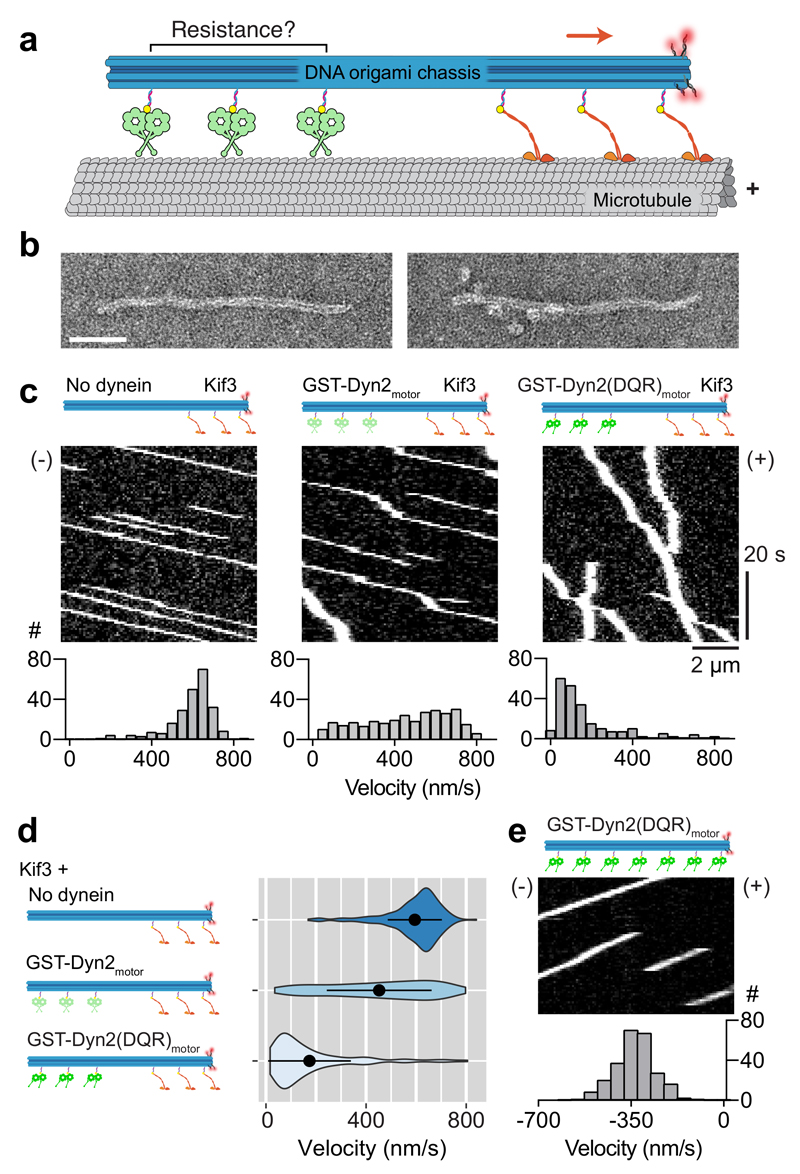
Figure 6. Assembly and motility of dynein-2 and kinesin Kif3 in multi-motor arrays.
(a) Diagram of the experimental set-up. A DNA origami chassis physically links arrays of IFT motors (in this example 3 Kif3 and 3 dynein-2). The movement of the resulting assembly on the microtubule is visualized via tetramethylrhodamine-labeled DNAs (red spheres).
(b) Negative stain EM images of DNA origami chassis samples with 3 dynein-2 sites before (left) and after (right) incubation with GST-Dyn2motor. Scale bar; 50 nm.
(c) Kymographs of chassis with indicated types of motor. (+) and (-) indicate microtubule polarity. Velocity histograms are shown below, in which the Y axis represents number of events (#). Number of runs analyzed: Kif3 chassis (231), GST-Dyn2motor-Kif3 chassis (299), GST-Dyn2(DQR)motor-Kif3 chassis (222).
(d) Velocity distributions for chassis type depicted as violin plots, with mean ± SD shown. Mean ± SEM values: Kif3 chassis (594.6 ± 7.1 nm/s), GST-Dyn2motor-Kif3 chassis (452.3 ± 12.1 nm/s), GST-Dyn2(DQR)motor-Kif3 chassis (173.8 ± 11.1 nm/s).
(e) Kymograph of chassis with 7 GST-Dyn2(DQR)motor sites and velocity histogram below (N = 249). Negative velocity indicates movement toward the microtubule minus end. See also Supplementary Fig. 6. Source data for c-e are available online.