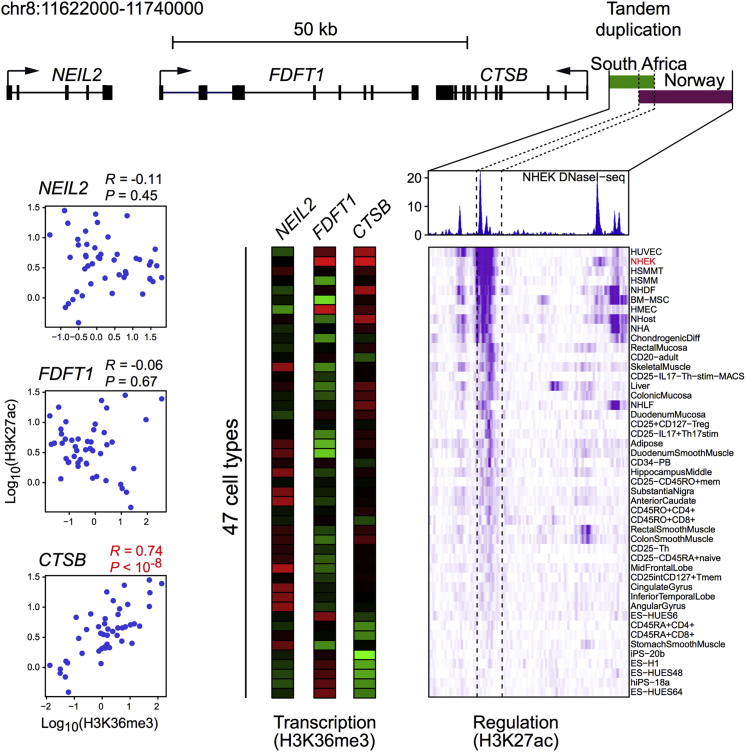
Figure 5.
Correlation of Enhancer Activity and Transcription of Nearby Genes in Cell Lines Reported by the ENCODE Consortium
The region of the South African (7.67 kb, green bar) and Norwegian (15.93 kb, purple bar) tandem duplications is displayed (a scale is shown at the top). The enhancer within the overlapping region was identified on the basis of a DNaseI hypersensitivity assay performed in NHEKs (blue peaks).28 Beneath this panel, the histone modification H3K27ac levels (ChIP-seq data) across the enhancer region in 47 human tissue and cell types are shown as a regulation heatmap (purple). Darker shades of purple indicate higher signal of the histone marker (H3K27ac), correlated with enhancer activity in the different tissues. Transcription of three nearby genes (CTSB, FDFT1, and NEIL2) was predicted by H3K36me3 levels (ChIP-seq data) across the genes in the same 47 different cell types. The transcription data are displayed as three parallel vertical bars in which red denotes high transcription activity and green denotes low activity. The correlations between H3K27ac and H3K36me3 levels are shown in the plots to the left. There was a highly significant positive correlation between the enhancer’s activity and CTSB expression (R = 0.74, p < 10−8).