Figure 2.
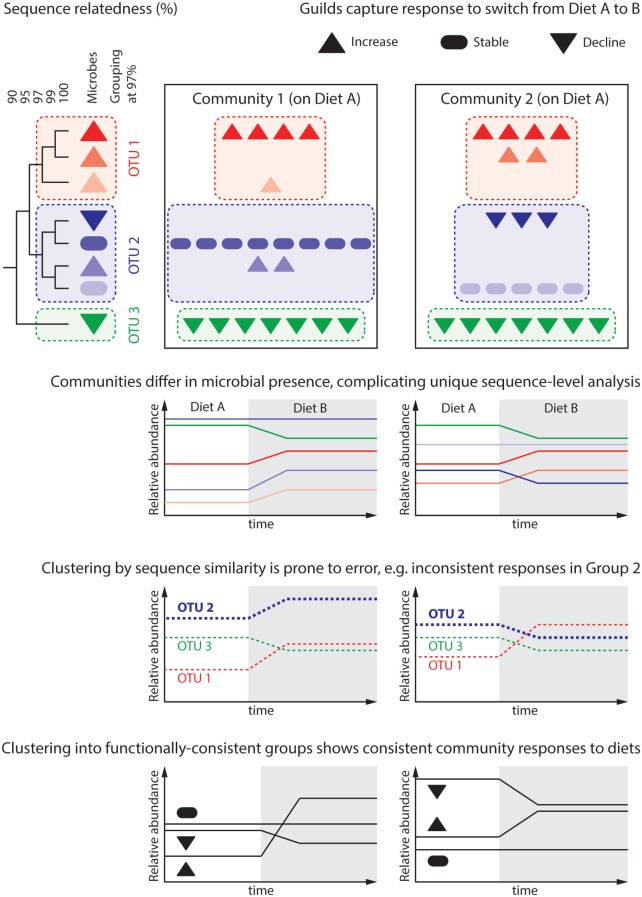
Predicting the microbiome response to dietary intervention requires that we define “units of response” capturing groups of microbes similarly responding to environmental parameters. The dendrogram depicts the genomic similarity of eight hypothetical microbes, which are differentially present in two communities. These communities are subject to a change in host diet, under which each microbe population either increases, declines, or remains stable. Attempts to reason about the community at the level of unique sequences are cumbersome as each community contains microbes that the other does not. This represents an over-classification, wherein several “units” (unique sequences) capture each possible response. Common practice in gut microbial ecology is to cluster unique sequences sharing 97% of their genomes into distinct groups, often termed “operational taxonomic units” (OTU). While analysis at the level of groups common to all communities can be performed, differential group constituent presence can still generate misleading results; this is because microbial traits, such as response to a given dietary change, do not correlate well with genomic similarity. For instance, Group 2 is seen to increase in Community 1, but decrease in Community 2. More meaningful is to group microbes by their response to environmental variables, in this case response to changing dietary intake: “guilds.” An open challenge for the microbial ecology field is predicting a microbe’s response to environmental variables based on either its genome or pure culture experiments.