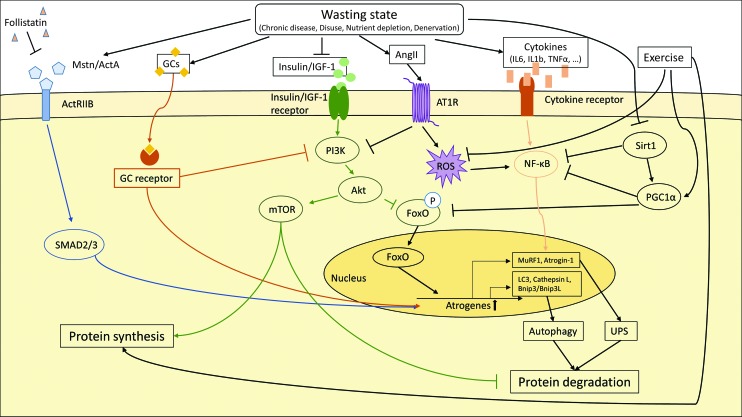
FIG. 5.
Molecular pathways involved in skeletal muscle protein degradation. The induction of atrogenes in catabolic muscle is driven by various molecular pathways. Upregulation of atrogenes leads to skeletal muscle protein degradation via the ubiquitin–proteasome system (UPS) and autophagy. Specific transcription factors such as FoxO proteins, NF-κB and SMAD2/3, as well as GCs activate the transcription of atrogenes. The transcription factors themselves are activated by external stimuli–myostatin (Mstn), activin A (ActA), GCs, insulin, IGF-1, and cytokines. In addition, the anabolic PI3K–AKT–mTOR activity is suppressed, which decreases skeletal muscle protein synthesis and leads to accelerated skeletal muscle protein degradation. Exercise, on the other hand, promotes protein synthesis, blocks ROS production, and enhances PGC1α gene expression. PI3K, phosphoinositide 3; AKT, V-Akt Murine Thymoma Viral Oncogene Homolog 1; mTor, mammalian target of rapamycin; MuRF1, Muscle RING Finger 1; Sirt1, NAD-dependent protein deacetylase sirtuin 1; IL1b/IL6, interleukin 1b/6; TNFα, tumor necrosis factor α; LC3, microtubule-associated protein 1 light chain 3; Bnip3/Bnip3l, BCL2 interacting protein 3/ligand; AT1R, Angiotensin II receptor type 1; ROS, reactive oxygen species; ActRIIb, activin receptor IIb; FoxO, forkhead box protein O; GCs, glucocorticoids; NF-κB, nuclear factor kappa B; PGC1α, peroxisome proliferator-activated receptor gamma coactivator 1-alpha. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars