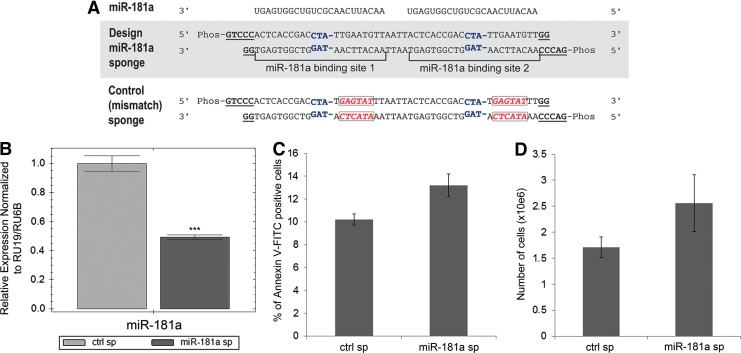
FIG. 6.
The effects of an miR-181a sponge inhibitor on IM-resistant K562-STI-R cells. (A) Schematic representation of the miR-181a sponge and the control sponge. The central mismatch at positions 11–14 (in which the nucleotide at position 14 was deleted) in each miRNA-binding site is shown in blue. The scrambled sequence for the control sponge is shown in red. Overhangs compatible with the restriction endonuclease SanDI are shown in underlined, bold font. (B) RT-PCR analyses of the levels of miR-181a in K562-STI-R cells electroporated with the control vector or that encoding the miR-181a sponge inhibitor. Expression relative to RU19 and RU6B is shown as fold difference. Triplicate analyses representative of data from three independent experiments are shown. ***P < 0.001 (C) FACS analyses of the percentage of apoptotic, Annexin V-FITC-positive K562-STI-R cells electroporated with the control vector or the miR-181a sponge inhibitor. Data represent four independent experiments. (D) K562-STI-R cells electroporated with the control vector or the miR-181a sponge inhibitor were cultured for 24 h. Cell count was determined. Data from three independent experiments are presented.