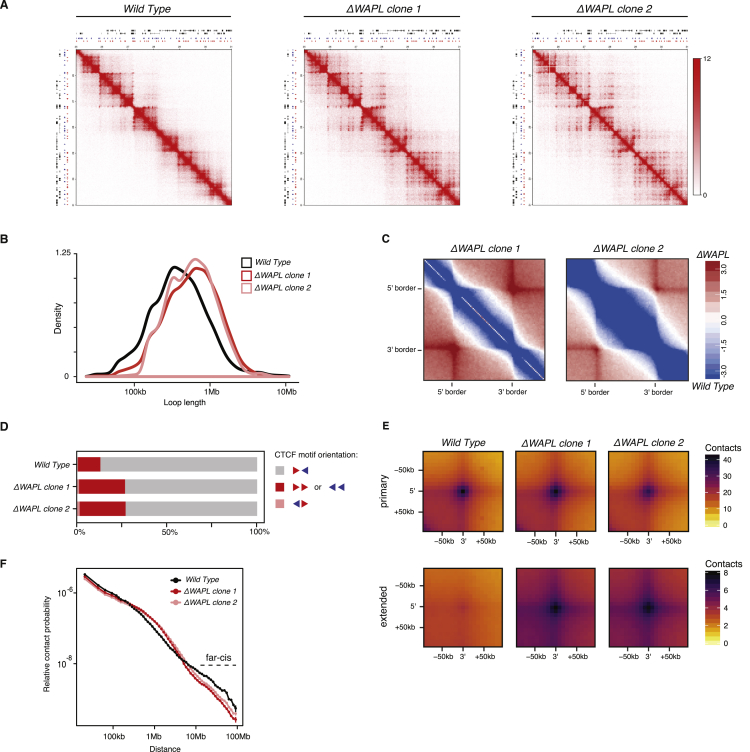
Figure S2.
The ΔWAPL Phenotype Is Also Observed in the Biological Replicate, Related to Figures 1, 2, and 5
(A) Hi-C contact matrices for a zoomed in region on chromosome 7 for wild-type, ΔWAPL clone 1 and ΔWAPL clone 2. Contact matrices are normalized to 100 million contacts, shown resolution is 20kb. Above and to the left of the contact matrices the union of CTCF sites identified in wild-type and ΔWAPL are shown. Red en blue triangles denote forward and reverse CTCF sites, respectively.
(B) Density plot showing the length distribution of the loops called by HICCUPS (Rao et al., 2014) in wild-type and ΔWAPL clone 1 and ΔWAPL clone 2.
(C) The differential ATA signal between both biological replicates of ΔWAPL and wild-type is visualized for all TADs. Blue indicates a higher signal in the wild-type, red indicates a higher signal in ΔWAPL cells.
(D) Quantification of the unique orientation of CTCF sites that could be associated with Hi-C loops called by HICCUPs.
(E) APA for primary and extended loops.
(F) Relative contact probability plot shows the likelihood of a contact at increasing length scales. Error bars depict the standard error of the mean.