Significance
A method that renders cells resistant to virus infection is reported. The method is based on anchoring antibodies to cell surface virus receptors to the plasma membrane. Because of their effective molarity, the antibodies are much more effective than if they were free in the circulation. The antibodies interact with the receptors and block virus attachment. In infections with viruses such as HIV, cells rendered resistant to infection will be selected over infected cells, thereby opening the possibility of curing the infection.
Keywords: HIV, cell surfaces, prevention
Abstract
Modern immunochemical engineering allows the creation of cells that either secrete antibodies or incorporate them into various cellular compartments, including the plasma membrane. Because the receptors for most viruses are known, if one can achieve the proper stoichiometry and geometry, plasma membrane-associated antibodies to these receptors should block viral infection. In this report, we test this concept for two different viruses, human rhinovirus and HIV. Plasma membrane-tethered antibodies efficiently rendered cells permanently nonpermissive for infection by both these viruses. Membrane-bound antibodies were much more efficient than free antibody in preventing infection, likely because of the effective molarity of membrane bound antibodies. Such resistant cells may restore immune-competence to otherwise compromised HIV patients.
The association of Ig with the plasma membrane of naive B cells creates the B-cell receptor (BCR) that interacts with its cognate antigen to initiate proliferation and differentiation of naïve cells into plasma and memory cells. The BCR also promotes specific antigen internalization to initiate antigen processing into peptides for presentation to helper T cells. Recently, we have constructed combinatorial antibody libraries expressing what we refer to as membrane-tethered antibodies (MTA) where all members of a large library are anchored to the plasma membrane (1, 2). In the MTA format, each cell in a culture has approximately 1 of 10 (8) different antibodies on its surface. When the same cell expresses an antibody and receptor, the system becomes autocrine and can be used to select for antibodies that bind to and either activate or inhibit the receptor. We have selected many such antibodies (2, 3), and the overall system appears to be general in that plasma membrane-bound antibodies and the receptor as an antigen always interact, likely because the effective molarity is so high.
We reasoned that this system could be used to construct cells where a plasma membrane-bound Ig binds to a cell surface receptor that doubles as a virus receptor to interfere with the latter’s ability to bind virus, thereby rendering the cells nonpermissive to infection. Further, because one is working with a large library of antibodies, it should be possible to select those that block virus interaction with the receptor while preserving the receptor’s physiological function.
Here, we report on the study of two systems, human rhinovirus and HIV. In the rhinovirus system, we learn the general principles for protection from infection by a highly aggressive pathogenic human virus and then apply these teachings to the medical need of constructing cells resistant to HIV infection where the immunochemical engineering of cell surfaces is most likely to find practical application.
Results
Rhinovirus.
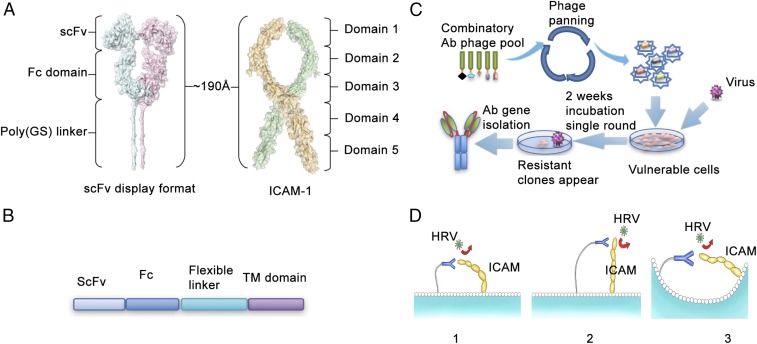
Ultimately, we wished to study the possibility that membrane-bound antibodies could completely protect the host cell from virus infection for both enveloped and nonenveloped viruses. We first studied rhinovirus as our model system because it is quite simple and robust and is easily handled without biosafety concerns. We targeted the ICAM protein because it is known to be the rhinovirus A and B receptor (4). Because the structure of the ICAM protein in complex with membrane-tethered antibodies is not known, we used a computer simulation to predict the optimum geometry for the interaction of the two proteins (Fig. 1A). The total length of the antibody plus linker (Fig. 1B) is approximately 190 Å, which is similar to the simulated full length of the ICAM protein. According to their size, three possible interaction models were proposed. They could both flex and move toward each other, or if ICAM itself is too rigid to bend, the antibody could bend to achieve contact because the linker is flexible. Alternatively, they could be brought into proximity by membrane curvature in the microdomains or trafficking vesicles (Fig. 1C). Because rhinovirus uses domain 1 located on the top of the receptor for docking, one should be able to select a functional anti-ICAM from combinatorial antibody libraries by using protection from virus induced cell death as the selectable phenotype (5).
Fig. 1.
Schematic representation of selection of Rhinovirus blocking, ICAM-binding antibodies. (A) Predicted structure and dimension of ICAM-1 ectodomain and the membrane tethered antibody. (B) The construct used to express antibody on the cell surface. (C) Phage displayed followed by the functional selection to identify the HRV-blocking antibodies. (D) Three models of proposed interactions between ICAM and membrane-tethered antibodies.
Screening for ICAM-1 Binding Antibodies That Prevent Infection.
We initially selected antibodies that simply bind to the ICAM protein by using a combinatorial human antibody library in phage with a diversity of >109 members. After two rounds of panning against the ICAM protein, the antibody encoding genes from the recovered phage particles were transferred to lentivirus vectors that were designed to deliver the gene to mammalian cells and express the antibody protein tethered to the plasma membrane (Fig. 1 B and D). The lentiviral vectors harboring the selected antibody encoding fragments were used to package a pool of lentivirus as the input library, and HeLa cells were infected at a MOI = 2 for the functional selection. The ability of lentivirus to express membrane-tethered proteins in this system was confirmed by using the florescence tomato protein in the place of the antibody fragment (Fig. 2A). The key finding is that the membrane-tethered protein covers the entire cell surface, indicating that when the MTA is expressed no trajectory for successful viral infection remains. To give more quantitative information, we used Western blotting versus antibody standards to estimate the number of MTA molecules linked to the plasma membrane of HeLa cells. There are approximately 30,000 MTAs expressed on the surface of each cell.
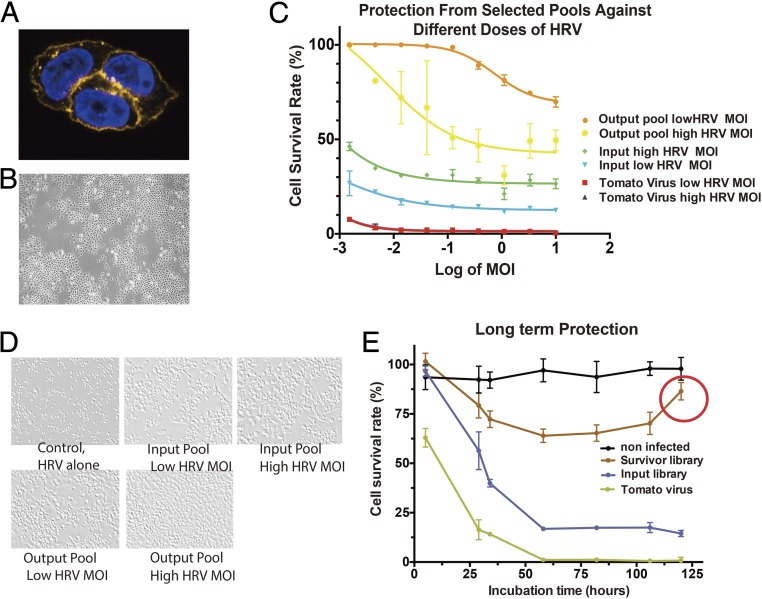
Fig. 2.
Selection of the HRV-blocking, ICAM-binding antibodies. (A) Staining of protein expressed in membrane-tethered format. (B) After 2 wk, dead cells and single colonies are present. (C) Kinetics of cell death. (D) Prevention of cell death by different pools of antibodies. “Input pool” stands for the lentiviral antibody sublibrary from phage panning, it is used as the first-round library for membrane-tethered lentivirus selection, which contains the antibody sequences from survivor cells. (E) Long-term protection.
Two days after infection with the pool of antibodies in lentivirus, rhinovirus was added to the HeLa cell culture at MOI = 1. The vast majority of cells died after 2 d as is normally seen. However, some virus-resistant colonies started to form and expand even in the presence of a high density of rhinovirus particles (Fig. 2B). Importantly, these studies showed that a selection based on cell survival can yield cells that are completely protected from cell death, presumably because of the MTAs linked to their surface.
All of the resistant colonies were collected for preparation of genomic DNA. The antibody-encoding fragments were recovered by PCR and then cloned again in the lentiviral vector. Direct comparison between the antibodies selected because they protect from infection (output pool) and the antibodies that were only selected for binding (input pool) showed that the output pool was much more effective in preventing infection. As a control, it was shown that lentivirus that only expressed tomato did not protect. (Fig. 2 C and D).
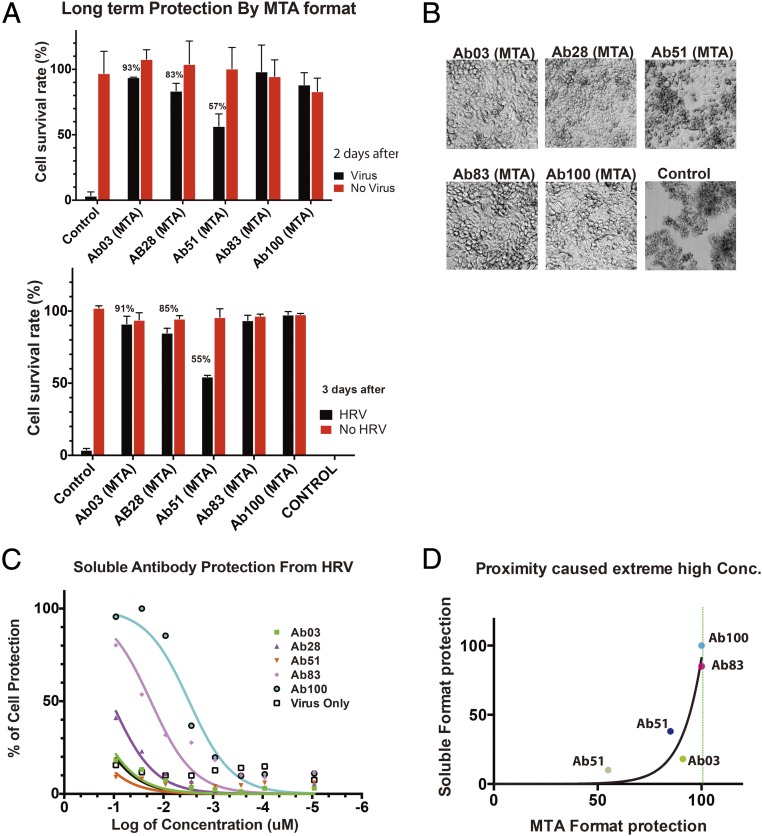
To generate individual antibodies, 100 bacterial clones were picked from the output library and sequenced. They were individually packaged into lentivirus, delivered to HeLa cells, expressed on the surface, and tested for protection from infection. Five different antibodies were identified, Ab03, Ab28, Ab51, Ab83, and Ab100. When examined 2 and 3 d after rhinovirus infection, all of them showed persistent protection, although to slightly different extents (Fig. 3 A and B). Interestingly, when expressed in the soluble format, the five antibodies demonstrated different abilities to inhibit rhinovirus infection (Fig. 3 C and D), with Ab100 as the strongest and Ab51 the weakest. Among different antibodies, the relationship between the degrees of protection by each soluble antibody at the highest concentration tested compared with the same antibody in the MTA format follows an exponential function, indicating the important role of effective molarity for the MTAs (Fig. 3E). Thus, the protective effect of a given antibody is vastly improved when it is used in the MTA format. It is important to note that the number of cells bearing MTA after an initial decrease in number actually increase over time, ultimately reaching the virtually 100% survival seen for uninfected cells (Fig. 2E, red circle). Because this increase in cell number occurs in the presence of virus, the initial killing followed by emergence of resistance has all of the features of a process based on selection.
Fig. 3.
Protection by individual antibody clones. (A) Protection by individual clones in MTA format. (B) Representative images of cells protected by isolated clones in MTA virus format. (C) Protection by different soluble antibodies. (D) Correlation of protection by purified antibody and by antibodies in membrane-tethered format. Different colored dots stand for different antibody clones. For each antibody, the percentage of protection in soluble format is plotted versus the percentage of protection in MTA format. The data points fits an exponential growth function with a τ = 9.7.
Selecting Antibodies That Preserve Normal Receptor Function by Paratope Fine-Tuning.
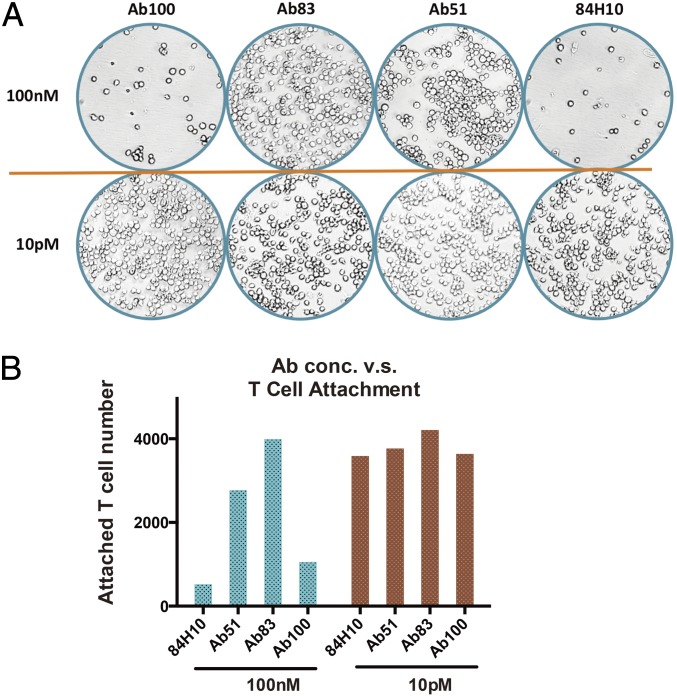
ICAM is known to mediate T-cell binding to endothelial cells by forming a complex with LFA. Because ICAM binding to LFA and rhinovirus use ICAM domain 1, there is a possibility that the rhinovirus blocking antibodies may interfere with the normal physiological functions of ICAM. This interference has been observed in other virus systems. For example, many of the CD4-binding antibodies block HIV entry efficiently but also comprise immune function that limits their clinical utility. Thus, we wished to test whether our antibodies inhibit normal ICAM1 function. A T-cell attachment assay was conducted as previously described except that we used high-content imaging to get a more accurate absolute count of the interacting T cells. At high concentration, the antibody that is most potent in protecting from virus infection (Ab100) is also the most efficient in blocking T-cell attachment, whereas other antibodies have more variable effects on T-cell binding (Fig. 4 A and B).
Fig. 4.
Antibody clones have different effect in blocking T cells attachment. (A) Representative images (acquired by GE6000 high content imager) of attached T cells at dish bottom. Top row is at higher antibody concentration (100 nM); the lower row is at lower concentration (10 pM). (B) Analysis of cell number attached to the dish bottom.
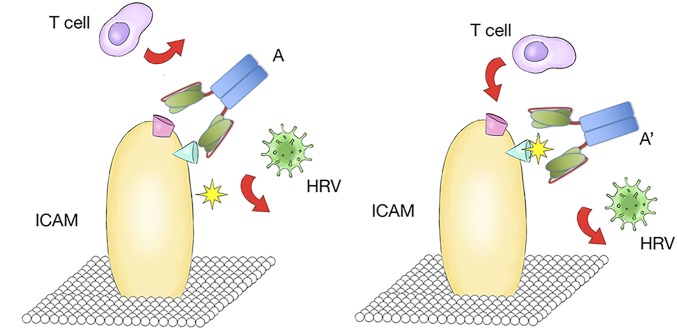
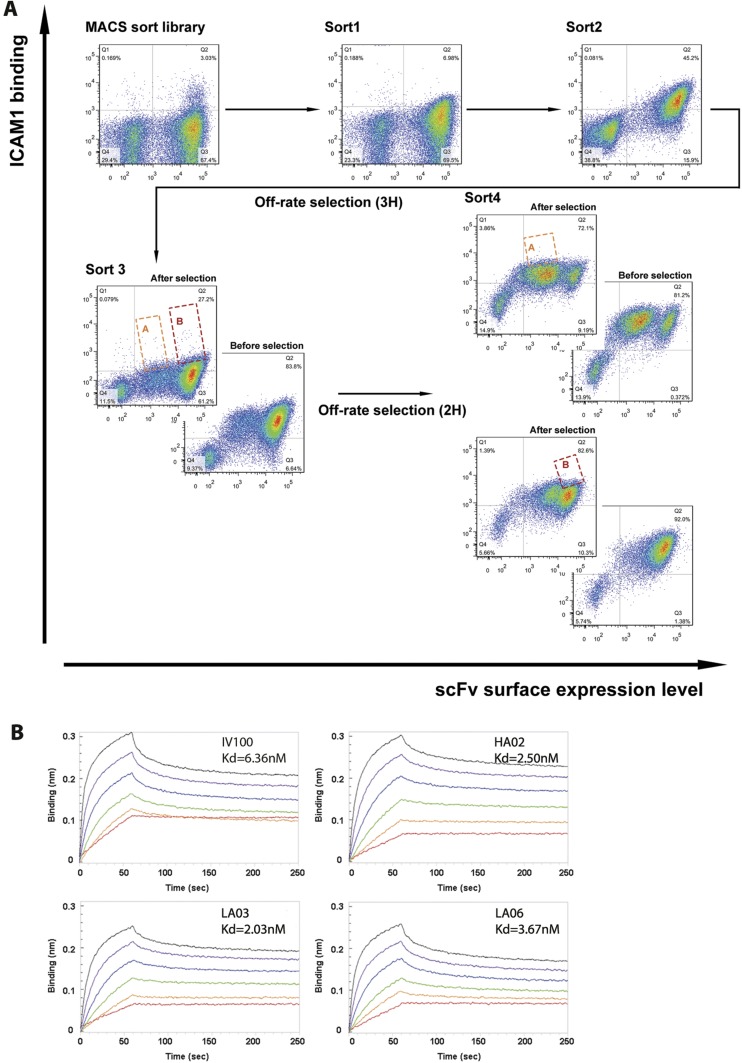
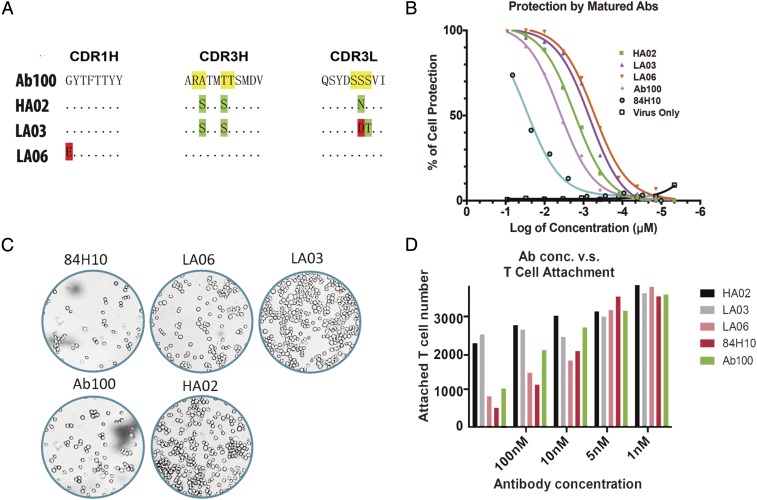
We thought that adjusting the paratope of the ICAM-1 binding antibodies might fine-tune the recognition mode, thus influencing retention of normal ICAM function (Fig. S1). Hence, we selected germ-line hotspots in Ab100 for generating a library of mutant antibodies. Because CDRH3 generally contributes more to antigen binding than CDR1 or CDR2, only hotspot residues in CDR3 of both light and heavy chains were chosen for mutagenesis. The library was designed to encode 10%∼50% of the wild-type amino acids. The other 19 amino acids had as many mutations as possible at each targeted position by the use of degenerate codons. We generated a library with a diversity of 2 × 108 in a yeast surface expression vector, pCTcon2. The screening procedure resembles standard yeast display, combining magnet-assisted cell sorting (MACS) and five rounds of flow cytometry-assisted cell sorting (FACS) to select for antibodies with the highest affinity (Fig. S2A). After five rounds of selection, 20 clones were picked from the output yeast pool and 3 were selected for further study based on their unique sequences (Fig. 5A). The KD for binding to ICAM by the newly selected antibodies was, 2.50 nM, 2.03 nM, and 3.67 nM, respectively for HA02, LA03, and LA06 compared with 6.36 nM for the original Ab100 (Fig. S2B). The antibodies were subsequently assayed for their ability to protect in vitro against rhinovirus infection. 84H10, a control antibody that is known to block ICAM function was determined to have an IC50 of 9 nM, whereas Ab100 has an IC50 of 2.4 nM. The mutant antibodies’ IC50 for blocking infection are 1.7 nM, 0.7 nM, and 0.5 nM for HA02, LA03, and LA06, respectively (Fig. 5B).
Fig. S1.
Schematic illustration of the paratope fine-tuning process. Altered recognition moved away from the T-cell binding sites to ICAM while still blocking rhinovirus entry.
Fig. S2.
Yeast display to select Ab100 mutant binders to ICAM. (A) FACS analysis of the ICAM antigen binding to the yeast pool. The different images represent different rounds of selection. After round 3, the pool was divided into subpool “A” and “B”, for further selection. (B) Measuring binding constants at different concentration of ligand.
Fig. 5.
Paratope fine-tuning adjusts the antibodies’ function. (A) The mutations identified in the selected antibodies from yeast display output. (B) Protection from rhinovirus infection by purified and modified versions of Ab100. (C) Representative images acquired by the high content imager. (D) Ab100 and its derivatives block ICAM-mediated T-cell attachment with different efficiency. 84H10 is used as a negative control because it is known to block T-cell attachment.
The T cells’ attachment assay of these mutant antibodies using analysis by high-content imaging showed that HA02 and LA03 at high concentration only blocked T-cell attachment by 30%, whereas blockage by LA06 was nearly complete (Fig. 5 C and D). Thus, it appears that one can select antibodies that retain their ability to neutralize virus infectivity, whereas preserving the normal function of the target receptor. For example, antibody from clone 83 has an IC50 for prevention of virus infection of approximately 10 nM, whereas it completely spares T-cell attachment at 100 nM.
Blocking HIV Infection by Membrane-Tethered Antibodies.
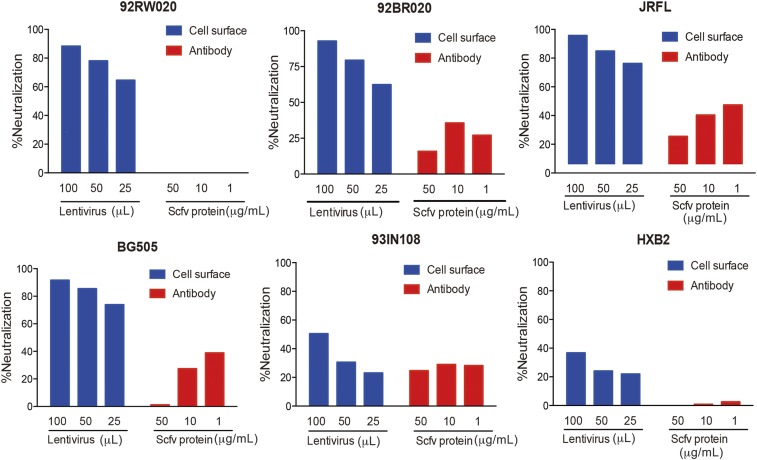
Encouraged by the results in the rhinovirus system, we turned to HIV. The idea was to determine whether MTAs against known HIV cellular receptors could render the cells resistant to infection. To test this concept, we studied anti-CD4 antibody, whichis known to be a receptor for HIV. We engineered TZM-bl cells to express anti-CD4 ScFv antibodies in a MTA format. These cells are widely used to evaluate antibody neutralization in standardized HIV-1 infectivity assays because the degree of virus neutralization in this assay correlates well with protection in nonhuman primates. The anti-CD4 antibody leu3a (converted to scFv) in a MTA format demonstrated almost complete inhibition of infectivity for four of six viruses (92RW020, 92BR020, JRFL, BG505) in a cross-clade virus panel and significant protection for the remaining two viruses (94IN108 and HXB2). Again, by comparison, the soluble version of the antibody was either nonneutralizing or poorly neutralizing even at a high concentration of 50 μg/mL (Fig. 6). The results of these studies again show that expression of antibodies on the cell surface of target cells results in enhanced neutralization activity compared with soluble antibody.
Fig. 6.
TZM-bl neutralization assays show that the α-CD4 antibody “Leu3a” was capable of preventing pseudovirus infection when expressed on the cell surface (blue) than when delivered as soluble antibodies (red). Antibodies were tested on a cross-clade 6 virus panel consisting of isolates 92RW020 (clade A), 92BR020 (clade B), JRFL (clade B), BG505 (clade A), 93IN108 (clade C), and HXB2 (clade B). Neutralization activities were dose-dependent on the amount of lentivirus added (100 μL, 50 μL, or 25 μL) and on the amount of soluble antibody added (50 μg/mL, 10 μg/mL, and 1 μg/mL).
Finally, in terms of stoichiometry, the number of expected MTA molecules per cell is in the range of the number of known HIV receptors on T cells (6). The absolute concentration of receptors appears to be critical for HIV infection. For example, for CCR5, when the receptor threshold falls below 10,000 molecules per cell, the ability to infect cells falls dramatically (7). Thus, MTAs can function by simply lowering the number of virus receptors below a critical density.
Discussion
In this study, we show that plasma membrane-tethered antibodies to virus receptors can inhibit entry of two prototype viruses, the nonenveloped rhinovirus and the larger enveloped HIV. Although antibody engineering of cell surfaces appears to be a promising approach to generating cells that have the potential to render the host resistant to virus infection, attention will inevitably turn to the most desirable properties of the antibody. This selection of antibodies happens in all cases where one ponders the use of antibodies to contain an infectious process whether it is by common vaccination or the special case of cell engineering as studied here. In our case, the most desirable antibody should have high affinity for the receptor and not interfere with the normal physiological function of the receptor. Toward this end, a major advantage of our approach is that it is not limited to receptors without critical functions because antibodies can be selected that block virus infection while sparing the native function of the receptor. Although such antibodies might be rare, nowadays we have technologies to select such rare antibodies from vast numbers of candidates in combinatorial libraries. Also, one might simultaneously target multiple receptors and coreceptors. The choice of the target receptor is, of course, important with CCR5, CD4, and CXCR4 being obvious choices that could be used either individually or in combination. Importantly, the efficiency of protection need not be absolute because one relies on the process of selection when the resistant cells are returned to the host. Thus, the T cells that are completely resistant to infection should expand because they have a growth advantage and could thereby lead to sustained resistance to HIV infection. Initially one expects the host to be a chimera of resistant and nonresistant cells, but over time the pool of resistant cells should become the majority.
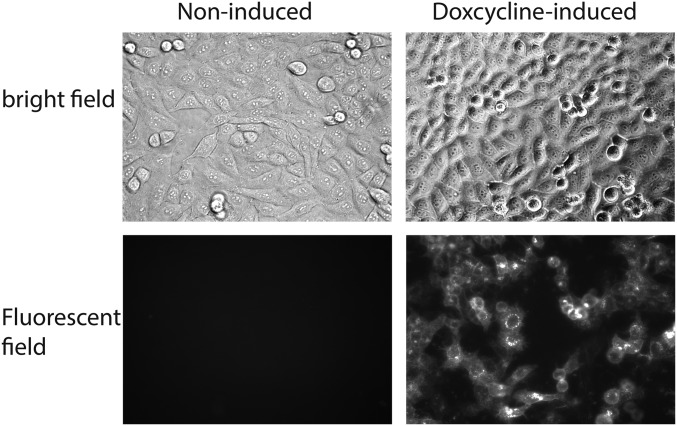
Thus, while we have proof of concept, the exact antibodies studied here are unlikely to be the ones ultimately used in clinical trials. This selection of antibodies is similar to what has already happened in the field with anti-gp120 antibodies because the community continues to discover ever better antibodies both in terms of potency and range of neutralization (8). Finally, we may wish to make the antibody gene inducible so as to be able to generate resistant cells at will. Indeed, when the fluorescent protein tomato is integrated into the lentiviral MTA construct under the control of a tetracycline-inducible promoter, the fluorescent signal on the surface is absent but can be switched on by adding doxycycline to the culture media (Fig. S3). Thus, the next steps toward a therapeutic regimen will involve selection of the best antibody in terms of specificity and potency followed by studies in primates.
Fig. S3.
Inducible promoter-driven Tomato-MTA expression upon doxcycline induction. The fluorescent protein tomato coding fragment was cloned into the LVX-tetOne (Clontech) vector, transfected to HeLa cells, and induced by 1 μg/mL doxcycline for 2 d.
There are, of course, other approaches to the generation of cells resistant to HIV infection. One widely discussed approach is the use of gene editing to eliminate integrated virus(s) or virus receptors. When it comes to integrated viral genomes, a major problem with gene editing is that the editing process accelerates the generation of replication-competent viral escape mutants that are now resistant to Cas9/sgRNA, thereby greatly limiting its potential for HIV therapy (9). Because our approach is not genetic, viral escape would have to involve development of an alternative route for virus entry into cells that is unlikely and, if it happened, would be expected to be highly inefficient and selected against at the population level.
Because patients with null mutations in CCR5 (CCR5 Δ32) may be resistant to HIV infection, another approach is to use gene editing to disable the CCR5 receptor (10, 11). However, the long-term consequences of having CCR5 defective alleles is not known, particularly when it comes to the importance of this chemokine receptor in combating other virus infections (12). This potential need to maintain the natural function of the receptor is one reason that we may wish to have inducible gene expression of the MTA so as to allow restoration of the receptor in certain clinical situations. Also, in some cases, chronically infected patients who received CCR5 Δ32 stem cell transplants suffered virus outbreak because of selection in vivo of CXCR4-tropic viruses (13). Again, this need for the receptor is why we may wish to use more than one MTA, particularly inclusive of an antibody against CXCR4 that allows retention of its critical physiological function.
Finally, it is of interest to place our work in perspective relative to approaches using vaccination, passive antibodies, or small molecule drug therapy. All these approaches are aimed at HIV itself, whereas MTAs target the host and work in conjunction with the process of selection. Basically, the goal comes down to the initiation of a process of real-time Darwinian selection in the host. Paradoxically, the virus becomes the driving force for selection of a resistant host who no longer harbors virus. For this selection to happen, one may need intervals such as “drug holidays” where viral replication is allowed, thereby using the power of selection to help eliminate reservoirs of infected cells.
Materials and Methods
See SI Materials and Methods for detailed description.
Construction of Full-Length ICAM-1 and ScFv-Linker Structure Model.
The structural model of MTA was built-in coot [PMID: 20383002] by using the scFv domain from PDB ID code 1P4I [PMID: 14754898] and the Fc domain from PDB ID code 1H3X [PMID: 12527303]. The 18-residue GS linker was added to the N-terminal of the Fc domain in an extended conformation. The structural model of ICAM was from PDB ID code 1Z7Z [PMID: 16004874].
Selection of Antibodies Against Human ICAM-1.
Recombinant human ICAM-1 ectodomain (Sino Biological) was biotinylated with EZ-link Sulfo-NHS-SS-Biotin (Thermo Fisher). The protein was used after removal of excess biotin reagent with a Zeba spin column (Thermo Fisher). The combinatorial antibody library in phage was incubated with the biotinylated ICAM-1 protein for 1 h. Streptavidin Dynabeads M-280 were then added into the mixture, and bound phage was eluted from Dynabeads M-280 by using glycine·HCl (pH 2.2). ICAM-1 binding phage were propagated by infecting XL1-blue cells. After two rounds of screening, the pooled scFv antibody library was subcloned to a lentiviral expression vector.
SI Materials and Methods
Construction of Full-Length ICAM-1 and ScFv-Linker Structure Model.
The structural model of MTA was built in coot [PMID: 20383002] by using the scFv domain from PDB ID code 1P4I [PMID: 14754898] and the Fc domain from PDB ID code 1H3X [PMID: 12527303]. The 18-residue GS linker was added to the N-terminal of the Fc domain in an extended conformation. The structural model of ICAM was from PDB ID code 1Z7Z [PMID: 16004874].
Selection of Antibodies Against Human ICAM-1.
Recombinant human ICAM-1 ectodomain (Sino Biological) was biotinylated with EZ-link Sulfo-NHS-SS-Biotin (Thermo Fisher). The protein was used after removal of excess biotin reagent with a Zeba spin column (Thermo Fisher). The combinatorial antibody library in phage was incubated with the biotinylated ICAM-1 protein for 1 h. Streptavidin Dynabeads M-280 were then added into the mixture and bound phage was eluted from Dynabeads M-280 by using glycine·HCl (pH 2.2). ICAM-1 binding phage were propagated by infecting XL1-blue cells. After two rounds of screening, the pooled scFv antibody library was subcloned to a lentiviral expression vector.
Production of Lentivirus in HEK 293T.
Lentivirus was produced by reverse transfection of 3.6 × 106 HEK 293T cells in a six-well plate, using 1.5 μg of lentiviral expression vector from the pooled anti-ICAM1 antibodies, 0.6 μg of pVSV-G and 0.8 μg of pCMV-dR8.9, and 7.5 μL of Lipofectamine 3000 (Thermo Fisher). After incubation at 37 °C for 6 h, medium was replaced with fresh DMEM with 10% FBS (D10F). Forty-eight hours after medium exchange, cell supernatant was centrifuged at 931 × g for 10 min at room temperature and collected. Cellular debris was further removed by a 45-μm pore filter. Lenti-X p24 rapid titer kit (Clontech) was used to determine the p24 level of lentivirus so that host cells can be transduced at a specific MOI. The virus preparations were aliquoted and frozen at −80 °C.
Phenotypic Selection Using the Selected Antibody Pool.
After one round of selection based on cell survival, genomic DNA from the surviving HeLa cells was recovered and used as a PCR template. The scFv fragment was amplified by PCR and subcloned into pFUSE-hIgG1-Fc vector (InvivoGen) for sequencing and identification of target gene enrichment.
Expression and Purification of the scFv-Fc Fusion.
Overrepresented scFv sequences in pFUSE-hIgG1-Fc expression vector were transfected into HEK 293F for expression. Four days after transient transfection of HEK 293F with the expression plasmids, supernatants were harvested and the scFv-Fc fusion proteins were purified by using a prepacked HiTrap Protein G HP column with ÄKTA avant 150 (GE Healthcare). Buffer exchange to PBS was performed by ultrafiltration. The purified antibodies were quantified by BCA assay and stored at 4 °C.
In Vitro Protection Against Rhinovirus Induced Cell Death Using a MTS Assay.
HeLa cells in DMEM were seeded at 2 × 104 cells per well in a 96-well plate to a final volume of 100 μL. After overnight incubation at 37 °C, the culture medium was replaced with D10F containing rhinovirus (MOI = 1) and serial concentrations of selected antibodies for 36 h at 30 °C. Twenty microliters of MTS solution (Promega) was then added to each well and absorbance at 490 nm was measured after 2 h incubation, following manufacturer’s instructions.
Mutant Library Construction for Affinity Maturation.
Germ-line hotpots were targeted for mutagenesis, as described (14). In short, RGYW/AGY mutational hotspots in CDR3 of clone IV100 were randomized by degenerate PCR mutagenesis. A mutant library was constructed by in vivo homologous recombination between the PCR products and linearized yeast display vector pCTCon2 (15). The library was transformed into the yeast strain EBY100 and grown in SD-CAA overnight at 30 °C. Following induction in SGR-CAA at 20 °C for 20 h with a starting OD600 ∼ 1, the yeast library was subjected to one round of MACS and five rounds of FACS.
MACS and FACS.
MACS was used for the primary library to reduce library diversity. The galactose-induced yeast cells were harvested and incubated with 50 nM of biotinylated human ICAM-1 at room temperature (RT) for 1 h. After washing and incubation with Streptavidin MicroBeads (Miltenyi Biotec) at RT for 30 min, ICAM-1 binding yeast-display scFv were then enriched by magnetic sorting over an LS column (Miltenyi Biotec). Recovered yeast were expanded in SD-CAA overnight at 30 °C.
Round 1–2 of FACS screening were performed by incubation of 2.5 nM biotinylated ICAM-1 with scFv-displaying yeast for 1 h until equilibrium binding was reached. Chicken anti-c-Myc IgY (Thermo Fisher) and Alexa Fluor 488 Goat Anti-Chicken IgG (H+L) (Thermo Fisher) were used to determine scFv expression on the yeast cell surface. Anti-Biotin-APC (Miltenyi Biotec), DyLight 633 NeutrAvidin (Thermo Fisher) were applied respectively in each round for the assessment of antigen binding.
Biotinylated ICAM-1 (0.5 nM) was used for incubation for round 3–5 of FACS screening, with the use of R-Phycoerythrin Streptavidin (Thermo Fisher), DyLight 633 NeutrAvidin, and R-Phycoerythrin Streptavidin in each round. In round 4 and round 5, selection pressure was increased by off-rate selection. In short, after incubation with biotinylated ICAM-1 and buffer wash, nonbiotinylated ICAM-1 was added at a final concentration of 100 nM into the scFv-displaying yeast and incubated for 2 h to outcompete biotinylated ICAM-1 from weak binders. Cells were acquired on a BD FACSAria III (BD Biosciences), and data were analyzed by using FlowJo (Treestar).
Isolation and Sequencing of Plasmids from R6 FACS Output.
Twenty randomly picked clones recovered from SD-CAA plates stemming from each gate of FACS round 5 were grown in YPD overnight at 30 °C. The yeast cells were harvested, and plasmid DNA was recovered by using yeast plasmid miniprep I (Zymo Research). Before sequencing, isolated DNA mixture were transformed into TOP10F for miniprep.
Kinetic Characterization of Antigen Binding Using Biolayer Interferometry.
Binding kinetics of the selected antibodies against ICAM-1 were performed on an Octet Red96 system (ForteBio) at 30 °C. Extracellular domain of ICAM-1 was biotinylated by using EZ-Link sulfo-NHS-LC-biotin (Thermo Fisher), and captured by streptavidin-coated biosensors (ForteBio). A series of the purified antibodies were titrated against the immobilized ICAM-1. Kilodaltons for each antibody were calculated applying a 2:1 interaction model.
PMA Jurkat Cell Adhesion Assay.
Human ICAM-1-Fc was coated onto a 96-well half-area high binding plate at 10 μg/mL PBS overnight at 4 °C. Plates were washed and blocked with 1% BSA in HBSS buffer for 1 h at RT. Antibodies were serially diluted with HBSS buffer containing 1% BSA and 2 mM MgCl2. The antibody solution was added to the plate (25 μL per well) and incubated for 15 min at RT. To each well was then added 25 μL of HBSS buffer containing 50 ng/mL PMA, 1% BSA, and 2 mM MgCl2. After 30 min, Jurkat cells were added to the plate (105 cells per well) in 100 μL of HBSS, 1% BSA and 2 mM MgCl2, and incubated at 37 °C/5% CO2 for 3 h. Cells attached on each well of the plate were imaged and quantified by using in Cell Analyzer 6000 following aspiration of the plate and washing with 200 μL of HBSS/2 mM MgCl2.
Pseudovirus Production and Neutralization Assays.
To produce pseudoviruses, plasmids encoding Env were cotransfected with an Env-deficient genomic backbone plasmid (pSG3∆Env) in a 1:2 ratio with the transfection reagent FuGENE 6 (Promega). Pseudoviruses were harvested 72 h after transfection for use in neutralization assays. Neutralizing activity was assessed by using a modified single round of replication pseudovirus assay and TZM-bl target cells, as described (16). Briefly, TZM-bl cells were seeded in a 96-well flat bottom plate at a concentration of 10,000 cells per well. For cell surface antibody display, the TZM-bl cells were infected with lentivirus for 48 h at 37 °C. For soluble antibody experiments, antibody was added to TZM-bl cells at the indicated concentrations and also incubated for 48 h. Pseudovirus was then added to these cells and incubated for an additional 48 h. Luminescence was then quantified 48 h following HIV pseudovirus infection via lysis and addition of Bright-GloTM Luciferase substrate (Promega).
Acknowledgments
We thank Dr. Rajesh Grover for helpful discussions.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702764114/-/DCSupplemental.
References
- 1.Lerner RA. Combinatorial antibody libraries: New advances, new immunological insights. Nat Rev Immunol. 2016;16:498–508. doi: 10.1038/nri.2016.67. [DOI] [PubMed] [Google Scholar]
- 2.Xie J, Zhang H, Yea K, Lerner RA. Autocrine signaling based selection of combinatorial antibodies that transdifferentiate human stem cells. Proc Natl Acad Sci USA. 2013;110:8099–8104. doi: 10.1073/pnas.1306263110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H, et al. Selecting agonists from single cells infected with combinatorial antibody libraries. Chem Biol. 2013;20:734–741. doi: 10.1016/j.chembiol.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Greve JM, et al. The major human rhinovirus receptor is ICAM-1. Cell. 1989;56:839–847. doi: 10.1016/0092-8674(89)90688-0. [DOI] [PubMed] [Google Scholar]
- 5.Xie J, et al. Prevention of cell death by antibodies selected from intracellular combinatorial libraries. Chem Biol. 2014;21:274–283. doi: 10.1016/j.chembiol.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci USA. 1999;96:5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reynes J, et al. CD4+ T cell surface CCR5 density as a determining factor of virus load in persons infected with human immunodeficiency virus type 1. J Infect Dis. 2000;181:927–932. doi: 10.1086/315315. [DOI] [PubMed] [Google Scholar]
- 8.Burton DR, Hangartner L. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu Rev Immunol. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, et al. CRISPR/Cas9-derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Reports. 2016;15:481–489. doi: 10.1016/j.celrep.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 10.Allers K, et al. Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation. Blood. 2011;117:2791–2799. doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- 11.Hütter G, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 12.Glass WG, et al. CCR5 deficiency increases risk of symptomatic West Nile virus infection. J Exp Med. 2006;203:35–40. doi: 10.1084/jem.20051970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kordelas L, et al. Essen HIV AlloSCT Group Shift of HIV tropism in stem-cell transplantation with CCR5 Delta32 mutation. N Engl J Med. 2014;371:880–882. doi: 10.1056/NEJMc1405805. [DOI] [PubMed] [Google Scholar]
- 14.Ho M, Pastan I. In vitro antibody affinity maturation targeting germline hotspots. Methods Mol Biol. 2009;525:293–308– xiv. doi: 10.1007/978-1-59745-554-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chao G, et al. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 16.Li M, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]