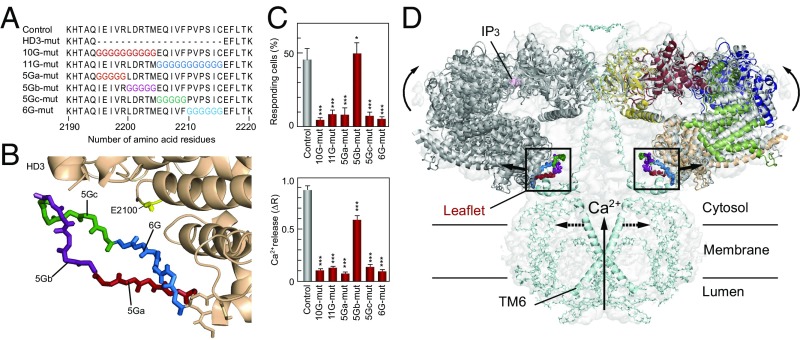
Fig. 5.
Critical sites in HD3 responsible for channel gating. (A) Amino acid sequences represent our mutated sites in which 10–11 amino acid residues (10G- and 11G-mut) or 5–6 amino acid residues (5Ga-, 5Gb-, 5Gc-, and 6G-mut) were substituted for glycine residues. (B) Mutated sites in the IP3R2217 crystal structure: 5Ga-mut, red; 5Gb-mut, purple; 5Gc-mut, green; and 6G-mut, blue. A critical glutamate residue, E2100, is shown in yellow. (C) Bar charts summarizing the relative population ± SEM (%) of 0.3 nM BK responding cells calculated from data of more than seven independent experiments (Left) and the amplitude of [Ca2+]i increase ± SEM (ΔR) calculated from data of more than 50 cells (Right) (control, n = 167; D2550A, n = 58; 10G-mut, n = 137; 11G-mut, n = 90; 5Ga-mut, n = 171; 5Gb-mut, n = 144; 5Gc-mut, n = 163; 6G-mut, n = 149). The significance of multiple comparisons was estimated by Dunnett’s method. *P = 0.0139; ***P < 0.001. (D) Model for IP3-dependent gating mechanism of IP3R. Two opposing IP3R2217 molecules are delineated by superimposing in the cryo-EM map (EMD-6369). Color codes of domains and critical sites in the leaflet structure are the same as in Figs. 1 and 3. The Ca2+ conducting pore formed with the transmembrane helix and the C-terminal region (light blue) were prepared according to coordinates of rat IP3R1 (PDB ID code 3JAV). We propose that IP3-dependent global conformational changes and gating transmission by the leaflet structure work in concert to open the channel pore (dotted lines).