Table 2.
Relative protein content changes in C. humilis leaves under water deficit
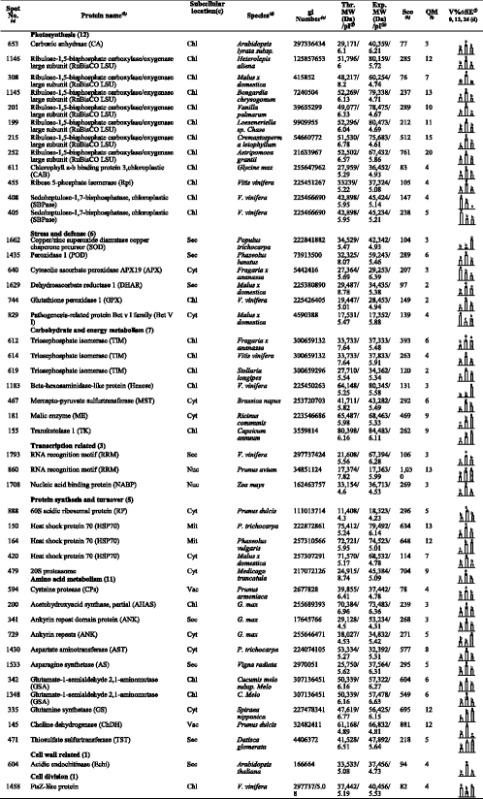
aAssigned spot number as indicated in Fig. 4. bThe name and functional categories of the proteins using MALDI TOF-TOF MS. cProtein subcellular localization predicted by softwares (YLoc, LocTree3, Plant-mPLoc, ngLOC, and TargetP). Only the consistent predictions from at least two tools were accepted as a confident result listed in Additional file 3: Table S3. Chl, chloroplast; Cyt, cytoplasm; Mit, mitochondria; Nuc, nucleus; sec, secreted; vac, vacuole.dThe plant species that the peptides matched. eDatabase accession numbers from NCBInr. f, gTheoretical (e) and experimental (f) mass (kDa) and pI of identified proteins. Experimental values were calculated using Image Master 2D Platinum Software. Theoretical values were retrieved from the protein database. hThe Mascot score obtained after searching against the NCBInr database. iThe number of unique peptides identified for each protein. j The mean values of protein spot volumes relative to total volume of all the spots. Four water deficit (0 d, 12 d, and 24 d) were performed. Error bars indicate mean ± standard error (SE)
