Abstract
Gene expression is regulated extensively at the level of mRNA stability, localization, and translation. However, decoding functional RNA regulatory features remains a limitation to understanding post-transcriptional regulation in vivo. Here, we developed RNA Element Selection Assay (RESA), a method that selects RNA elements based on their activity in vivo and uses high-throughput sequencing to provide quantitative measurement of their regulatory function with near nucleotide resolution. We implemented RESA to identify sequence elements modulating mRNA stability during zebrafish embryogenesis. RESA provides a sensitive and quantitative measure of microRNA activity in vivo and also identifies novel regulatory sequences. To uncover specific sequence requirements within regulatory elements, we developed a bisulfite-mediated nucleotide conversion strategy for large-scale mutational analysis (RESA-bisulfite). Finally, we used the versatile RESA platform to map candidate protein-RNA interactions in vivo (RESA-CLIP). The RESA platform can be broadly applicable to uncover the regulatory features shaping gene expression and cellular function.
Introduction
Post-transcriptional control is critical to regulate cellular protein levels. Mutations disrupting mRNA regulatory pathways have been linked to developmental defects and human diseases1–4, underscoring the importance of regulatory mRNA elements. Post-transcriptional regulation occurs through mRNA processing, nuclear export, translation, localization and stability, all of which significantly impact gene expression. This control is achieved by specificity factors that typically recognize elements within mRNA 5′ and 3′ untranslated regions (UTRs).
Decoding mRNA regulation transcriptome-wide remains a challenge. Pioneering work assaying sequence fragments (‘bashing’) combined with mutational analysis has identified regulatory sequences within individual transcripts4–8, laying the groundwork for high-throughput methods that have expanded our understanding of mRNA regulation. RNA profiling approaches have illuminated the microRNA (miRNA) regulatory landscape by identifying miRNA targets transcriptome-wide9–12, refining miRNA-mRNA targeting rules12–14, and mapping miRNA binding sites on target mRNAs15,16. Additionally, alternative 3′UTR sequences promoting mRNA stability have been discovered by measuring isoform half-life differences17.
Recent approaches combine plasmid reporters with custom oligonucleotides to discover sequences with regulatory potential18–22. However, these methods have disadvantages for comprehensively identifying discrete regulatory elements across a transcriptome (Supplementary Table S1). Oligonucleotide libraries are a trade-off between sequence diversity and length. Short oligonucleotides22 cannot represent regulatory elements with longer sequence requirements, while longer random oligonucleotide libraries19 can only contain a subset of the exponentially large number of possible sequences, which may not adequately represent all biologically relevant elements. Targeted libraries that curate sequences according to criteria such as conservation18,21 are by design limited in scope, while approaches that build reporters from 3′UTRs in full23 or in large windows20 have limited resolution to identify discrete elements. Finally, plasmid delivery constrains library sequence diversity, and can require normalization during analysis.
To achieve a comprehensive map of the mRNA regulatory landscape, we have engineered an in vivo reporter system with flexible sequence input and the capacity to select functional RNA elements across large candidate regions, based on biological function. Here, we describe RNA Element Selection Assay (RESA), a high-throughput technique that measures regulatory activities of mRNA sequences with near nucleotide resolution. Rather than relying on custom oligonucleotide synthesis, RESA couples sequence amplification with random fragmentation to generate highly-overlapping fragments spanning RNA regions of interest. This library is introduced into an in vivo system, subjected to experimental treatment, then assayed for differential selection using RNA-Seq. This flexible framework has also enabled us to develop RESA-Bisulfite, which introduces mutations across the library to assay for regulatory nucleotide requirements, and RESA-CLIP to select for specific protein-RNA interactions.
We have applied these techniques to characterize determinants of mRNA stability during zebrafish embryogenesis. We find that RESA provides a sensitive and quantitative measure of miRNA activity in vivo and reveals sequence features that enhance miR-430-induced mRNA destabilization. RESA additionally reveals novel regulatory regions independent of miR-430. This method provides a valuable tool to enhance our understanding of how mRNA stability is regulated during development, and provides a framework to annotate RNA regulatory elements across a variety of biological contexts.
Results
RESA design and implementation
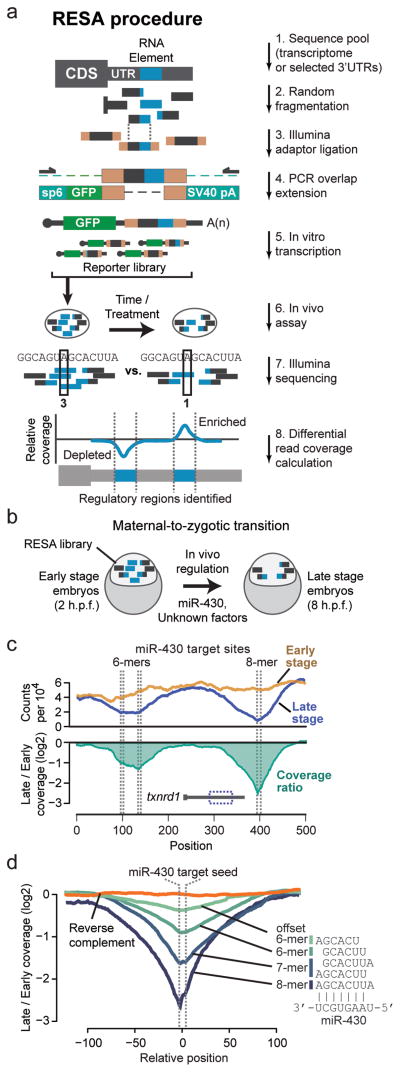
To systematically identify regulatory mRNA sequences in vivo, we developed RNA Element Selection Assay (RESA). This method measures the regulatory strength of diverse sequence fragments by quantifying position-specific changes in abundance using high-throughput sequencing (Fig. 1a). To generate the RESA library, we isolated 3′UTR sequences by PCR amplification, induced random fragmentation by sonication, ligated Illumina sequencing adaptors, and using overlap-extension PCR introduced them into the 3′UTR of a GFP open reading frame with SP6 promoter and SV40 polyadenylation signal. This strategy avoids cloning and generates a library of high sequence complexity. As a by-product of library preparation, reverse-complement inserts are also generated; these can serve as internal controls for their forward-direction counterparts (Supplementary Fig. 1a,b). Alternatively, this method can utilize existing RNA-Seq libraries as source DNA for the overlap-extension PCR, which enables interrogating regulatory activity across a transcriptome. In both cases, in vitro transcription generates a diverse mRNA reporter pool with identical 5′ UTR, CDS, and polyadenylation signal, but variable 3′UTR (Fig. 1a).
Figure 1. RESA is a high-throughput method to systematically map regulatory mRNA sequences in vivo.
(a) Pipeline for RESA library generation. The RESA library is in vitro transcribed from templates assembled from random sequence fragments, and introduced into an in vivo system. Positional coverage from Illumina sequencing using built-in adapters reveals regions of depletion or enrichment, as a read-out of regulatory activity (i.e. destabilizing or stabilizing). (b) RESA application to mRNA stability during the zebrafish maternal-to-zygotic transition, when maternal mRNAs are destabilized by miR-430 and unknown factors. (c) (top) Coverage profile for regions overlapping miR-430 target seed sequences (dashed lines) within the txnrd1 locus, at early stage (yellow) and late stage (blue). (bottom) Ratio of late versus early stage profiles (green). (d) Meta profiles of late vs early coverage ratios centered on the miR-430 target seed sequence, grouped by seed complementarity: 8-mers (N=8), 7-mers (N=31), 6-mers (N=94), offset 6-mers (N=95), and reverse complement 8- and 7-mers (N=39).
Embryonic development in all animals requires global remodeling of the transcriptome (reviewed in24), thus providing an ideal context to study post-transcriptional regulation. To interrogate mRNA regulation during the maternal-to-zygotic transition (MZT) in zebrafish, we built a RESA library of 3′UTR fragments from maternally-provided mRNAs (Fig. 1b), spanning 434 genes and 502,728 nucleotides (Supplementary Data 1,2).
Identifying regulatory RNA elements with high resolution requires extensive sampling of fragments throughout the mRNA. To assess sequence diversity, we sequenced the in vitro transcribed RESA library, using the built-in Illumina adapters. We found that sonication yielded a highly diverse population of overlapping fragments, 56 to 129nt, that mapped throughout the UTRs (Supplementary Fig. 1c). This diversity was maintained after injection into embryos (Supplementary Fig. 1d). 97% of the UTRs had ≥20X median sequencing coverage across their length in the injected library, while 87% had ≥100X coverage (Supplementary Fig. 1e). Unique read start sites occurred at 69% of all nucleotide positions in the library, and per UTR, a median 83% of nucleotides overlapped read start sites (Supplementary Fig. 1f), indicating a high abundance of unique fragments. Together, these results demonstrate that RESA efficiently generates an mRNA library of extensive sequence complexity.
RESA detects and quantifies discrete regulatory mRNA elements with high resolution
To assess how effectively RESA detects RNA regulatory potential in vivo, we injected the library into replicate sets of 1-cell embryos (Supplementary Table S2) and performed RNA-Seq at 2 hours (“early stage”) and 8 hours (“late stage”) post fertilization to identify fragments that become enriched or depleted as a measure of their regulatory activity in the cognate mRNA (Fig. 1b). We first evaluated RESA’s ability to identify miR-430-induced mRNA destabilization. miR-430 is zygotically transcribed3,25 and triggers translation repression, deadenylation and destabilization of several hundred maternal mRNAs, canonically by recognizing complementary target sequences (GCACUU) within target 3′UTRs3,9,26. The RESA library includes 29 validated miR-430 target 3′UTRs9. Whereas sequencing read abundance across the library was highly correlated between replicate timepoints (R ≥ 0.97) (Supplementary Fig. 1h,i), we observed strong valleys of depletion between different timepoints in fragments overlapping validated miR-430 target seeds, such as in the txnrd1 3′UTR9 (Fig. 1c, Supplementary Fig. 1g). The greatest magnitude of depletion coincided precisely with the nucleotide position of the target seeds (Fig. 1c), suggesting that RESA can specifically map the effect of miR-430 mediated destabilization.
The number of contiguous base pairs between the miRNA seed and target site correlates with targeting efficacy14,26. Accordingly, we found that the magnitude of fragment depletion was significantly stronger for greater matches to the miR-430 seed (8-mers > 7-mers > 6-mers > offset 6-mers) (P < 3.8E-39, Kruskal-Wallis test) (Fig. 1d, Supplementary Fig. 2a,b), demonstrating RESA’s ability to quantify the strength of in vivo regulation.
RESA identifies RNA elements regulating mRNA stability
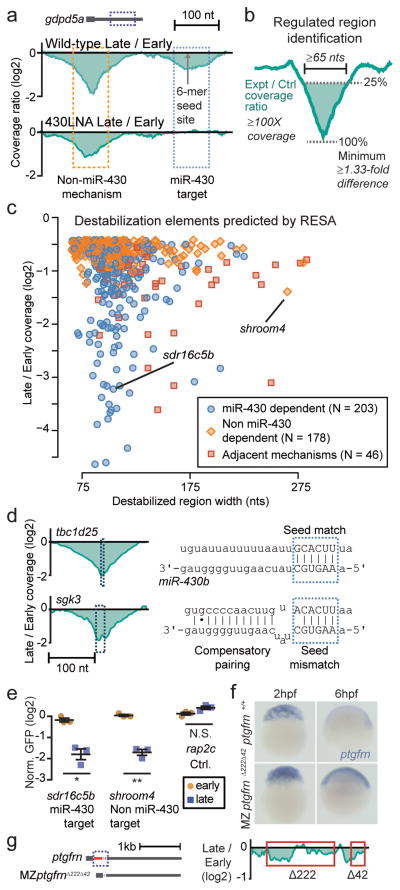
Next, we searched the entire RESA library for differentially regulated regions. To distinguish miR-430 dependent and independent effects, we repeated RESA in the absence of miR-430 function using a tiny locked nucleic acid complementary to the miR-430 seed (430LNA)27. Under these conditions we find that miR-430 mediated regulation is specifically blocked (Fig. 2a; Supplementary Fig. 2c,d), thus allowing us to isolate the effects of non miR-430-dependent destabilization.
Figure 2. RESA identifies sequences that promote mRNA destabilization.
(a) Wild-type sequence coverage depletion over time (top) reveals two destabilization elements in the gdpd5a 3′UTR. 430LNA inhibits destabilization of only the miR-430 target (bottom). (b) Regulated regions are identified by searching for a ≥ 1.33 fold depletion in late versus early stages, within a ≥65-nt region with ≥100X sequencing coverage. (c) Biplot of RESA-predicted destabilization regions, separated by mode of regulation. (d) Wild-type coverage depletion around a canonical miR-430 6-mer site (tbc1d25) and non-canonical seed mismatch site with 3′ compensatory pairing (sgk3) (e) GFP reporter analysis of RESA-predicted regulatory element with different regulatory behaviors. GFP mRNA levels are assayed at early (2hpf; yellow circles) and late (8hpf; blue squares) stage by qRT-PCR and normalized to a control dsRed mRNA. Plot shows mean +/− SEM; **, P<0.01; *, P<0.05, Welch’s t-test. (f) In situ hybridization to detect endogenous ptgfrn mRNA levels in maternal-zygotic mutants for the predicted destabilization element (MZptgfrnΔ222Δ42) compared to ptgfrn+/+ at 2 and 6h.p.f. (g) (left) Diagram of the predicted destabilization element in the ptgfrn 3′UTR (dashed box) and the CRISPR-mediated deletion (red) in MZptgfrnΔ222Δ42. (right) RESA coverage profile over the destabilized region.
To identify regulated regions, we parameterized a search strategy based on features of miR-430 targets. We performed simulations on a known miR-430 regulated mRNA to measure the effects of varying read coverage on target site discovery, and determined that we could confidently recover destabilized regions with ≥1.33-fold decrease in sequencing coverage from early to late stage, given ≥100X coverage (Supplementary Fig. 3). Using these parameters on pooled replicates, with a region width threshold of ≥65nt as measured at 25% of the valley minimum (Fig. 2b, Supplementary Fig. 4), we found 427 destabilized regions across 273 3′UTRs (Fig. 2c). Of these, 52% showed evidence of regulation in the absence of miR-430 function (Fig. 2c, Supplementary Fig. 5–6, Supplementary Data 3), including 46 regions that appear to be composed of adjacent miR-430-dependent and independent elements (Fig. 2c, Supplementary Fig. 5c). The majority of destabilized regions (86%) are robustly identified even using individual replicates in isolation (Supplementary Fig. 7a–c), and all 29 of the previously validated miR-430 targets9 are correctly classified, demonstrating the efficacy of this method. An additional 400 lower-confidence destabilizing elements were identified, as well as 361 regions that appear to enhance stability in late-stage embryos (Supplementary Fig. 6b, 7d, Supplementary Data 3), suggesting that multiple mechanisms regulate mRNA levels during embryogenesis.
miR-430 target identification was highly specific. 93% of 8-mer target sequences and 86% of 7-mers fell within high-confidence miR-430 dependent destabilized regions. Weaker seed matches were less likely to be destabilized, with 35% of offset 6-mer sequences showing no evidence of regulation, reflecting the existence of other features that modulate targeting efficacy14 (Supplementary Table 3). 82% of target seed-containing regions classified by RESA as miR-430 dependent have valley minima within 10nts of the seed (Supplementary Fig. 7e), reflecting the high nucleotide precision of predicting regulatory element location with RESA. In addition to canonical seed-based regulation, RESA identified 46 miR-430 dependent regions lacking a seed match (Fig. 2d, Supplementary Fig. 8a–b), suggesting the prevalence of non-canonical miR-430 targeting. These regions are highly enriched for partial, 5-nt matches to the miR-430 seed (P = 4.3e-13, Chi-squared test), as well as complementarity to the 3′ end of mature miR-430 (P = 7.3e-5, Wilcoxon rank sum test) (Fig. 2d, Supplementary Fig. 8c–e), consistent with previous observations that non-canonical target-miRNA base pairing can also induce destabilization1,14,28,29.
We independently verified the regulatory activity of RESA-identified regions by generating reporters and assaying mRNA levels in embryos by qRT-PCR. We observed reporter destabilization consistent with RESA predictions (Fig. 2e, Supplementary Fig. 6b), indicating that these regions are sufficient to confer destabilization in vivo. To test the necessity of RESA-identified regions for endogenous destabilization, we generated deletions of the RESA-predicted destabilizing regions in ptgfrn using CRISPR-Cas930(Fig. 2f–g). Maternal-Zygotic (MZ) mutants fail to clear ptgfrn mRNA (Fig. 2f), thus showing that the sequences identified by RESA are required for mRNA destabilization in vivo. All together, these results demonstrate that RESA effectively identifies regulatory elements that confer differential mRNA stability.
RESA-Bisulfite identifies nucleotide requirements in regulatory elements
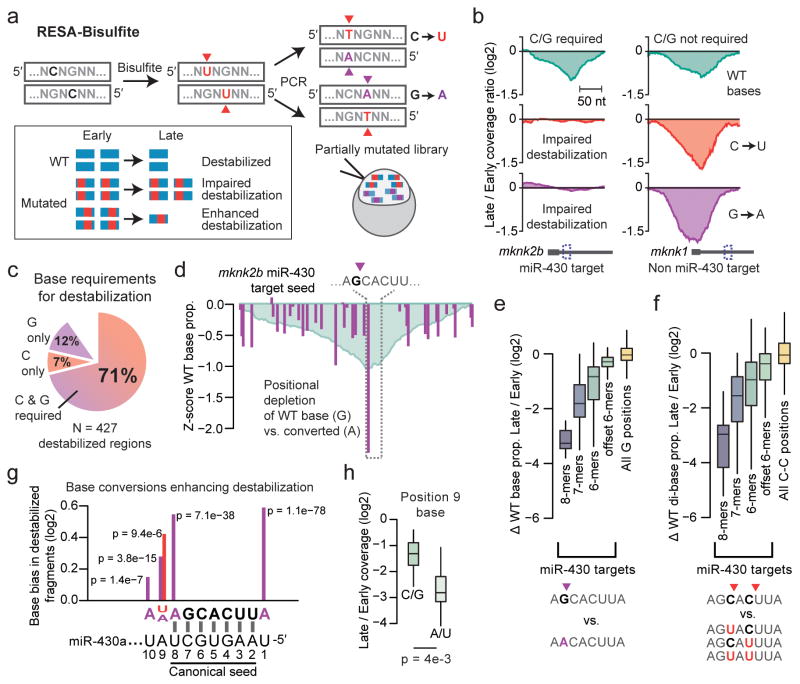
Elucidating specific sequence requirements driving regulatory element activity typically involves measuring the effects of mutations within the element. However, conventional targeted mutational analysis is a challenge for large numbers of elements. To address this, we developed an extension to RESA called RESA-Bisulfite (RESA-B), which introduces mutations across a RESA library to assay the effect of disrupting regulatory elements. Bisulfite treatment induces cytosine deamination at a proportion of sites. After PCR, each RESA insert encodes either C to T or G to A mutations in the forward orientation, at random positions (Fig. 3a), to generate a highly complex library of mutated sequences that can be assayed for activity and compared to non-mutated libraries.
Figure 3. RESA-Bisulfite implements high-throughput mutational analysis for regulatory sequences.
(a) RESA-Bisulfite used bisulfite treatment to induce C to U or G to A mutations within the reporter library pool. Partially mutated reporters are injected in vivo to determine the effect of mutations (red, purple) on the destabilization of wild type (WT, blue) regions. (b) Coverage ratio plots for reads with wild-type bases (top), C to U converted bases (middle), and G to A converted bases (bottom) for a region within mknk2b that requires C and G bases for destabilization (left) and a region within mknk1 that does not require C or G bases for destabilization (right). (c) Proportion of destabilized regions identified by RESA that require C and/or G bases for destabilization. (d) Positional analysis of G to A conversions over the mknk2b miR-430 target seed at late stage, showing positions with G depletion (purple). Y-axis measures the Z score of the ratio of WT base count to mutated base count at each position, compared to all positions. RESA signal is overlaid in blue. (e,f) Depletion of wild-type G bases versus converted A (e) and C-C dibases versus converted U-C, C-U, and U-U (f) in late-stage versus early-stage embryos across all miR-430 targets grouped by seed complementarity: 8-mers (N=13), 7-mers (N=40), 6-mers (N=136), offset 6-mers (N=115). All G (N=99157) and C-C (N=92696) positions across the library also shown. Boxes show median and quartiles, whiskers show 1.5 times the interquartile range. Outliers not shown. (g) Pooled base conversion biases adjacent to miR-430 target seeds showing enrichment of wild-type bases relative to converted. Y-axis shows the magnitude of change in the proportion of converted bases observed (U, red; A, purple) from early to late stage. X-axis is oriented sense to the target. P value from a G test of independence is shown. Axes as in (h). (j) Boxplots comparing RESA destabilization of miR-430 targets encoding endogenous A or U at position 9 (N=20) versus C or G (N=22) for 8-mers and 7-mers (pairing positions 2–8). Boxes show median and quartiles, whiskers show 1.5 times the interquartile range. Outliers not shown.
We created a RESA-B library from the same UTR fragments as the original RESA experiment, injected it into 1-cell embryos, and sequenced RNA extracted at 2 and 8 hours. After read alignment using a procedure to handle mutated bases (Supplementary Fig. 9), we observed a 68% conversion rate of C and G positions (Supplementary Fig. 10a–b), suggesting that a large proportion of fragments would contain mutated, and hence potentially ineffective, regulatory elements. Indeed, many destabilized regions presented reduced activity in RESA-B, suggesting that they require C and G bases (Fig. 3b,c).
To identify individual C/G positions required for regulatory element activity, we quantified the proportion of wild-type versus converted bases at each position. For discrete elements such as miR-430 target sites (GCACUU), single-base changes can affect function. In a mixed population of partially converted reporters, molecules encoding wild-type target sites would become destabilized and depleted, but those with converted sites, e.g. ACACUU, would be more stable and become over-represented (Fig. 3d). Indeed, G bases are significantly depleted in miR-430 target seeds over time, correlated with seed strength (P = 2.1e-95, Kruskal-Wallis test) (Fig. 3e), demonstrating that RESA-B can detect selective stabilization of miR-430 targets mutated at single nucleotides. Similarly, the proportion of unconverted C-C di-bases is significantly depressed in miR-430 target seeds (P = 3.5e-63, Kruskal-Wallis test) (Fig. 3f), reflecting their combined regulatory contribution (Online Methods). Across the entire library, we found 2060 positions with significantly depleted wild-type bases from early to late stage (individual G tests of independence, FDR < 0.05) (Supplementary Fig. 10d, Supplementary Data 4). Taken together, RESA-B facilitates large-scale mutational analyses that can reveal regulatory element base requirements.
RESA-Bisulfite identifies base changes that enhance regulation
Mutations can also create sites conferring stronger regulation (Fig. 3a, Supplementary Data 4). We found that offset 6-mer miR-430 targets adjacent to a wild-type C (AGCACUc) were significantly less depleted compared to mutated sequences that convert the target into a stronger 7-mer (AGCACUu) (P = 2.1e-20, G test of independence) (Supplementary Fig. 10e). We also identified base changes outside the miR-430 seed region that enhanced destabilization. Converted bases are strongly depleted at the non-seed positions 9 and 10 (Fig. 3g), suggesting that A and U strengthen miR-430 targeting. We confirmed this effect in the original RESA library (Fig. 3h, Supplementary Fig. 10f), and using reporters, we found that changing position 9 to A in an endogenous target increased destabilization by 35% (Supplementary Fig. 10g), demonstrating additional targeting determinants for miRNA-mediated regulation. In summary, RESA and RESA-B provide a framework to discover novel sequence attributes that contribute to mRNA regulation.
RESA-CLIP maps RNA-protein interactions
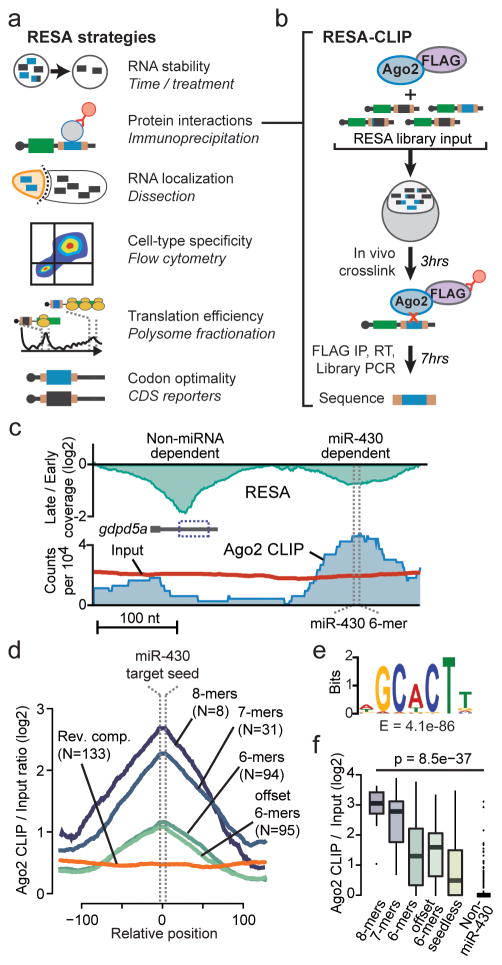
RESA allows identification of regulatory RNA activity by differential accumulation, and we envision that this approach can be extended beyond analysis of mRNA stability by combining RESA libraries with various selection strategies (Fig. 4a). To demonstrate one such application, we developed RESA-CLIP to map RNA-protein interactions (Fig. 4b). mRNA regulation is often mediated by specific trans-acting RNA-binding proteins (RBPs), whose target sequences can be identified using crosslinking and immunoprecipitation (CLIP-Seq). Present methods are labor intensive, require high sequencing depth, and involve challenging downstream computational analyses (Supplementary Fig. 11a). We reasoned that RESA could be an ideal template for quickly identifying RNA-protein interactions in vivo because of two key advantages. First, since the RESA library is pre-built with Illumina adaptors, RNA fragmentation and inefficient adaptor ligation are not necessary to generate the sequencing library. Second, the results can be analyzed with existing CLIP-Seq pipelines, or using the RESA framework.
Figure 4. The RESA framework can be adapted to map RNA-protein interactions and other RNA biology in vivo.
(a) Diagram of proposed examples of RESA applications in diverse biological contexts. (b) Overview of RESA-CLIP strategy. Tagged Ago2 is co-injected with the RESA library, injected into zebrafish embryos, crosslinked, immunoprecipitated, and sequenced to identify Ago2-bound regions. (c) RESA coverage ratio for the gdpd5a locus (top) and RESA Ago2 CLIP (blue region) and input library (red line) counts (bottom). RESA-verified miR-430 6-mer target site is marked with dashed lines. (d) Meta profiles of Ago2 CLIP versus input ratios centered on the miR-430 target seed sequence, grouped by seed complementarity and compared to reverse-complement inserts overlapping 8-, 7-, and 6-mers. (e) Motif analysis in Ago2 CLIP peaks shows significant enrichment of the miR-430 target seed sequence. (f) Box plots showing Ago2 CLIP enrichment over input across miR-430 dependent destabilized regions containing 8-mer target sites (N=11), 7-mers (N=33), 6-mers (N=74), offset 6-mers (N=24), and lacking a canonical target seed (N=61) compared to non miR-430 dependent regions (N=178). P values are from a Kruskal-Wallis test comparing all groups, and from a Wilcoxon rank sum test comparing seedless targets to non targets. Boxes show median and quartiles, whiskers show 1.5 times the interquartile range, points are outliers.
To perform RESA-CLIP, we co-injected the RESA library with FLAG-tagged Ago2 to assay binding to miR-430 targets (Fig. 4b). This entire procedure, from injection to sequencing library, requires only 10 hours, in contrast to other CLIP-seq protocols that require several days (Supplementary Fig. 11a). We observed an enrichment of Ago2 CLIP fragments overlapping miR-430 target sequences, corresponding to the strength of seed complementarity (Fig. 4c–e, Supplementary Fig. 11b).
At high stringency, 97% of CLIP peaks overlapped RESA-predicted miR-430-dependent destabilized regions, while at lower stringency, peaks are still highly specific for miR-430 regulated regions, with higher CLIP enrichment observed for stronger seed matches (P = 8.5e-37, Kruskal-Wallis test) (Fig 4f, Supplementary Fig. 11c–f, Supplementary Data 5). Notably, even RESA-predicted miR-430 dependent regions lacking complementary target seeds are significantly enriched for Ago2 binding over non-miR-430 target regions (P = 3.6e-7, Wilcoxon rank sum test) (Fig. 4f), demonstrating that these are likely bona fide miR-430 targets. Among the CLIP peaks not overlapping RESA-predicted destabilized regions, we did not find any significant sequence motifs indicative of other miRNAs; however, we did find extensive partial complementarity to miR-430 (Supplementary Fig. 11g), suggesting that the Ago2 CLIP indeed reflects the activity of only miR-430. Together these results show that the RESA platform can be extended to identify biologically relevant RNA-protein interactions in a rapid, high-throughput and quantitative manner.
Discussion
Here we describe RNA Element Selection Assay (RESA) to measure mRNA cis-regulatory element activity in vivo with near nucleotide resolution. We provide three applications to support potential widespread use of this method. First, we used RESA to determine sequences regulating stability in maternal mRNAs. Second, we developed RESA-Bisulfite to facilitate large-scale mutational analysis of cis-regulatory activity. Third, we demonstrated that RESA-CLIP efficiently maps RNA-protein interactions in vivo.
Sequence selection methods (SELEX)31–33 made it possible to determine sequence binding specificity in vitro. In vivo adaptations have relied on custom-made oligonucleotide libraries18–22, which can suffer from limited sequence complexity and resolution. RESA provides a versatile and quantitative platform to identify RNA regulatory activity in vivo, using a direct PCR assembly approach that maximizes molecular complexity of the query sequence pool. In vitro library transcription eliminates the need to normalize RNA levels to plasmid DNA quantities, which can vary significantly per cell. RESA enables the analysis of regulatory sequence potential in diverse organisms compatible with exogenous RNA introduction, including zebrafish, mouse, Xenopus, Drosophila, and C. elegans, and can also be adapted to cell culture with RNA transfection methods. RESA also streamlines sequencing library preparation by building in Illumina adapters, thus avoiding inefficient adaptor ligation and allowing robust library recovery. These innovations yield high sensitivity with broad applicability, and enable transcriptome-wide surveys of regulatory sequence activity.
We have also shown that RESA-CLIP can interrogate RNA-protein interactions without the cost and labor associated with other pull-down protocols, though because RESA-CLIP assesses protein binding with exogenously-supplied sequence fragments, detection is limited to elements that maintain activity outside of their native context. Despite this caveat, RESA-CLIP can provide a simple and rapid strategy to identify sequences that underlie binding specificities for many known RBPs.
RESA applies a selection principle based on differential accumulation of specific sequences to identify regulated regions, which can be easily applied to interrogate diverse levels of post-transcriptional control (Fig. 4a). For example, RESA can improve upon the identification of mRNA localization elements34–35, in combination with existing methods to isolate specific embryonic compartments, cell types, or subcellular structures. Similarly, polysome fractionation can distinguish highly and poorly translated mRNAs, allowing high-throughput identification of sequences directing translation activation or repression. Finally, RESA can be adapted to investigate how different codons36–38 or regulatory elements within coding regions39,40 shape mRNA stability and translation. We envision widespread usage of RESA variations to decode post-transcriptional mechanisms of gene expression across biological settings.
Online Methods
RESA reporter library construction
3′UTR regions of approximately 850–1300bp fragments were PCR amplified from 2hpf embryo cDNA with Taq polymerase for 36 cycles and annealing temperature of 55 °C. Gene definitions and corresponding gene symbols were obtained from Ensembl r79 on the zebrafish Zv9 genome (for figures, gdpd5a is gdpd5 and sgk3 is C24H8orf44). Primers were designed using Primer341 and are listed in (Supplementary Data 1). Half of each PCR reaction was pooled and purified on PCR cleanup columns (Qiagen #28104). To generate PCR library approximately 4μg DNA (50μL) was sheared using Covaris sonicator at full power for 20 min, ends were repaired, and Illumina adapters were ligated. Following adaptor ligation fragments were selected sizes ranging from 130–250bp to exclude adaptor dimer species.
Reporter library was generated by overlap extension PCR using Phusion High Fidelity DNA Polymerase (NEB #M0530) for 5 cycles of 98 °C, 61 °C, and 72 °C for 30 sec each. Then, 24 reactions using 2μL of elongated product was amplified for 10 cycles, annealing at 57 °C, with Forward GCTTGATTTAGGTGACACTATA and Reverse GAATTAAAAAACCTCCCACACC primers which map to the sp6 promoter and downstream of the SV40 polyadenylation signal. Reporter library template 1270bp was purified on 1% agarose, and 400ng was used for in vitro RNA synthesis using Sp6 Message Machine (ThermoFisher AM1340). Zebrafish embryos were injected with 10pg mRNA library and collected at the indicated stages.
miR-430 inhibition
To inhibit miR-430 function, zebrafish embryos were injected with 1nL of 10μM concentration of tiny locked nucleic acid complementary to the miR-430 seed (430LNA), 5′-TAGCACTT-3′ (Exiqon) as described in 27.
Sequencing library construction
Total RNA was extracted from 30–75 zebrafish embryos injected with the reporter library using Trizol reagent according to the manufacturer instructions. PolyA+ mRNA was selected with NEB magnetic beads (NEB S1419S). Briefly, 250μg or 50μL of beads were washed with 1X wash buffer (0.5M NaCl, 20mM Tris-HCL (pH 7.5), 1mM EDTA) for each sample. 3μg total RNA from each sample was diluted in 25μL water, incubated at 65 °C for 2 min to denature secondary structure and then placed on ice. 25μL 2X Wash/binding buffer (1M NaCl, 40mM Tris-HCL (pH7.5), 2mM EDTA) was added to each RNA sample, mixed with pre-cleaned beads, vortexed gently, and incubated at 40 °C for 10 min with occasional agitation. After 10 min binding reaction, supernatant was discarded and beads were washed twice with 100μL 1X wash buffer. Third wash was performed with 100μL ice-cold low salt buffer (0.15M NaCl, 20mM Tris-HCL (pH 7.5), 1mM EDTA). RNA was eluted with 25μL Elution buffer (10mM Tris-HCL (pH 7.5), 1mM EDTA) prewarmed to 65 °C for 2 min. Second elution was performed with the same conditions. Poly(A) selected RNA was recovered with Ethanol precipitation and resuspended in 15 μL of water. 6μL of RNA were reverse transcribed with reporter specific primer (CATCAATGTATCTTATCATGTCTGGATC) using SuperScript III. Sequencing library was prepared with 16 cycles of Fusion PCR with Forward primer-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTC and reverse CAAGCAGAAGACGGCATACGAGAT(barcode)GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT.
Computational and statistical analysis
All analyses were performed using custom scripts written in R and Python 3, except as noted below. All box plots show median, lower and upper quartile, and whiskers to 1.5 times the interquartile range; individual points represent outliers beyond the whiskers.
The following statistical tests were performed: Fig. 2e: two-sided Welch’s t-test on N = 3 independent sets of embryos (t = 6.0454, p = 0.015 for sdr16c5b; t = 11.7406, p = 0.0035 for shroom4; t = −2.7369, p = 0.053 for rap2c). Fig. 3e, f: Kruskal-Wallis rank sum test (statistic = 446.8683, p = 2.1e-95; statistic = 297.6536, p = 3.5e-63); Fig. 3g: individual G-tests for independence, results reported in Supplementary Source Data; Fig. 3h: two-sided Wilcoxon rank sum test (W = 333, p = 0.0038); Fig 4f: Kruskal-Wallis rank sum test (statistic = 179.0614, p = 8.5e-37) and two-sided Wilcoxon rank sum test (W = 3469.5, p-value = 3.6e-07). Supp. Fig. 2b,c: Kruskal-Wallis rank sum tests comparing targets to background (statistic = 186.0093, p = 3.8e-39 for Late/Early; statistic = 209.2651, p = 3.8e-44 for Late/430LNA); two-sided Wilcoxon signed rank tests (V = 2908.5, p = 7.2e-40 for Late/Early; V = 1604.5, p = 9.4e-45 for Late/430LNA); Supp. Fig. 8c: Chi-squared test (statistic = 52.4877, df = 1, p = 4.3e-13); Supp. Fig. 8d: two-sided Wilcoxon rank sum test (W = 19897, p = 7.3e-05); Supp. Fig 10e: G-tests for independence (G = 85.7295, df = 1, p = 2.1e-20 for C bias; G = 33.6585, df = 1, p = 6.6e-9 for G bias); Supp. Fig 10f: two-sided Wilcoxon rank sum tests (W = 2158.5, p = 0.0025 for offset 6-mers; W = 2613.5, p = 0.0017 for 6-mers; W = 333, p = 0.0038 for 7&8mers); Supp. Fig. 10g: two-sided Welch’s t-tests on N = 3 independent sets of embryos (t = 4.1776, p = 0.017 for pam A9 late vs early; t = 13.9267, p = 0.00066 for pam G9 late vs early; t = 6.9051, p = 0.0054 for itpr3 A9 late vs early; t = 35.9421, p = 4.2e-06 for itpr3 G9 late vs early; t = −5.2482, p = 0.027 for pam A9 late vs G9 late; t = −6.5417, p = 0.019 for itpr3 A9 late vs G9 late); Supp. Fig. 11e: two-sided Wilcoxon rank sum test (W = 723, p = 6.1e-08); Supp. Fig. 11f: Chi-squared test (statistic = 17.0887, df = 1, p = 3.6e-05); Supp. Fig. 11g: Chi-squared test (statistic = 18.7968, df = 1, p = 1.5e-05) and two-sided Wilcoxon rank sum test (W = 19941, p = 2.1e-09).
Sequencing read processing and alignment
Paired-end sequencing reads were pre-processed to trim any Illumina sequencing adapters from the 3′ ends42, then aligned to the zebrafish Zv9 genome using TopHat v2.0.1343, allowing only alignments consistent with the gene isoforms used to design the amplicons (Supplementary Data 2) by providing amplicon-specific transcriptome GTF definitions. The TopHat parameters used were --no-discordant --no-mixed --library-type fr-firststrand --transcriptome-only --read-mismatches 7 --read-gap-length 7 --read-edit-dist 7 --segment-mismatches 3 --b2-very-sensitive
Prior to alignment, the genome sequence was modified to maximize concordance with RESA library design: 1) all base positions outside of the amplicon regions were masked with “N”; 2) certain primer pairs used to amplify the library encode non-template restriction site overhangs; thus, to ensure maximal read alignment at the ends of these amplicons, the genome sequence was changed to match the overhanging primer sequence, for each such primer.
An alternative to this approach would be to simply extract the spliced transcript sequences for each UTR, exclusively align reads to a transcriptome index, then back-convert the transcriptome-relative coordinates to genome coordinates for downstream analysis.
Calculating read coverage
Positional sequencing coverage (i.e., pileup) was calculated individually for each UTR, such that each nucleotide position encodes the total number of sequence fragments that overlap it, using pooled biological replicates. In detail: first, the samtools version 1.244 view command was used to obtain all sequencing reads overlapping the genomic coordinates of a UTR. Each read pair was merged into a single sequencing read fragment spanning the start and end coordinates of the 5′ and 3′ read, respectively. Only read fragments with end-to-end lengths between 56 and 129 nts, inclusive, were used, based on the observed distribution of fragment lengths (Supplementary Fig. 1c,d), to avoid the effects of aberrant read alignments that would artificially produce shorter or longer genomic spans. Genomic coordinates were converted to UTR-relative coordinates. Read coverage was incremented by one at each base position between the start and end coordinates of the fragment, inclusive, only if the strand of the read fragment (defined by the strand of Read 1) was sense relative to the UTR. X coverage, relative to an experimental condition, is the raw count at a given nucleotide position. UTRs with 0 median sequencing coverage in the pooled early stage samples were considered to be failed PCRs and were excluded from further analysis. For antisense coverage (Supplementary Fig. 1a), only read fragments in the opposite strand orientation of the UTR are counted.
The log2 ratio of normalized coverage between two experimental conditions was used to identify position-specific differential regulation. In detail: First, raw counts in each experiment are normalized by dividing by the total count in that experiment (sum of all counts in all positions across all UTRs. Next, a smoothing factor (0.5) is added to each count to prevent zero division. Finally, the log2 ratio is calculated.
miR-430 target site analysis
Putative miR-430 target sites were identified using the Zv9 genome sequence. UTR sequence matches to AGCACTTA are considered 8-mers, AGCACTT and GCACTTA are 7-mers, GCACTT are 6-mers, and AGCACT are offset 6-mers. Seed mismatch sites are considered any 1-nt difference from the GCACTT 6-mer. 3′ complementarity was measured using Smith-Waterman local alignment to the reverse complement of each mature miR-430 sequence excluding the seed (430a: CTACCCCAACAAAT, 430b: CTACCCCAACTTAAT, 430c: CTACCCCAAAGAGA). Parameters were match=1, mismatch=0, gap=-100. The complementarity score is the number of dinucleotide matches in the best local alignment. Sequences used to estimate background frequencies were obtained from the UTR regions in the library not predicted to be regulated: for each such segment >= 65 nts, the middle 50 nts were used. For all population and meta analyses, only miR-430 target sites with at least 100X sequencing coverage in the pooled early stage libraries were used. Meta analyses additionally excluded target sites closer than 91 nucleotides to each other, to avoid measuring the overlapping influences of two independent targets; and excluded target sites too close to the ends of the UTRs, depending on the size of the region window being queried, to avoid artifacts due to truncation. To detect polymorphisms at miR-430 seeds in the libraries compared to the reference genome, samtools5 mpileup was run on the Early stage alignments followed by bcftools and vcftools commands to obtain the consensus sequence from the sequencing reads (samtools mpileup –uf Zv9.fa <bamfiles> | bcftools call -c | vcfutils.pl vcf2fq > utrs.fq).
Regulated region identification
Destabilized and stabilized regions were identified separately. For destabilized regions, positions at local minima of log2 coverage ratio were identified in each UTR as sites of destabilization. Broader regulated regions are defined as the minimal window around a local minimum such that each position in the window has a log2 coverage ratio within 0.25 times the local minimum. Stabilized regions are identified analogously using local maxima. In detail: using a sliding window of 100 nts across a UTR, the position of minimal log2 coverage ratio relative to all other positions in the window is recorded (local minimum). If this minimum value occurs at the last position of the window, suggesting an even lower value occurs downstream, no local minimum is called, and instead the window is shifted to the right by half a window length to continue searching. For each local minimum identified, the boundary of the regulated region is extended to the left and right, such that each position encountered has a log2 coverage ratio < 0.25 times the local minimum. Regulated regions that overlap are merged.
From this list of candidate destabilized regions, post hoc criteria are applied to classify the region as high confidence (width >= 65 nts, coverage at early stage >= 100X at the minimum, value <= −log2(4/3) at the minimum) or low confidence (width >= 40 nts, value <= −log2(5/4) at the minimum, no threshold for coverage). These parameters were selected based on the width of miR-430 associated regions (Supplementary Fig. 4a) and false-discovery rate simulations (below).
For each region, the sub-region most likely to contain the responsive sequence element was defined to be bounded by the left-most and right-most position with at least 0.9 times the local minimum (or maximum). For the purpose of motif analysis, this sub-region was extended 15 nts left and right, while not exceeding the boundaries of the regulated region.
Regulatory element width analysis
To identify the determinant of RESA region width, we reasoned that this property is the combined effect of regulatory element length requirements and sequencing fragment length. The characteristic “valleys” centered on miR-430 target seed sequences span 150–200nts, approximately twice the insert fragment length obtained during RESA library construction plus the 6–8 nts of miR-430 complementarity. To test the effect of sequence fragment length, aligned read pairs were restricted to short (56–80nts) or long (105–129nts) length classes based on the end-to-end distance of each read pair as reported by Tophat. RESA regulated regions were identified using both sets of reads individually and compared to RESA analysis using all reads (56–129nts) (Supplementary Fig. 4)
False-discovery rate estimation
To simulate the effects of sequencing coverage on false discovery rate, the acat1 UTR was selected to perform read sampling (Supplementary Fig. 3). At full coverage, acat1 encodes a single, well-defined miR-430 target site. For each simulated coverage level (e.g., 100X, 40X), a random subset of the overlapping reads was obtained for both early-stage and late-stage. log2 coverage ratio was calculated, and destabilized regions were predicted, with no threshold for size or magnitude. Among the regions (if any) not corresponding to the known miR-430 target site, the minimal log2 coverage ratio was recorded as the false-positive destabilization magnitude. This process was repeated 500 times per coverage depth, inducing a distribution of false-positive coverage ratios. At that coverage depth, an FDR of 0.1 corresponds to setting a threshold equal to the 10th percentile of the false-positive distribution.
Motif analysis
To identify overrepresented sequences in the regulated regions, the MEME Web tool v4.11.245 was used in discriminative mode. The control sequences consisted of all UTR regions in the library not predicted to be regulated (UTR regions minus regulated regions, discarding any segments <= 25nts). Default parameters were used, searching on the sense strand only with a target motif width of 6 to 12 nts.
RESA-bisulfite (RESA-B)
To preserve the integrity of sequencing adaptors 1ng RESA library was amplified for 10 cycles with primers containing methylated Cytosines, a modification that disfavors Cytosine deamination following bisulfite exposure. Library with hemi-methylated adapters (1.4ug) was deaminated using EZ DNA Methylation-Direct Kit (Zymo Research #D5020) according to manufacturer directions. The only modification to the protocol was to perform deamination reaction at 98 °C for 8 min followed by 64 °C for 25 min to reduce the deamination rate to ~50% according to a pilot assay of individual sequences using Topo cloning. Deaminated library was amplified for 7 cycles at 68 °C annealing temperature with non-methylated primers to fix mutations. A pool of 12 PCR reactions was Ethanol precipitated, separated on 8% PAGE, and 180–220bp region was purified. The mutated product was built into a GFP reporter library and injected into zebrafish as described above.
Analysis of bisulfite-treated libraries
Alignment strategy is outlined in Supplementary Fig. 9. Read pairs are first subjected to wild-type genome alignment to filter out fragments that cannot be unambiguously classified as C to T or G to A converted. Read pairs unaligned during this filtering step are then subjected to each of the four mutated alignment protocols in succession, with the expectation that only one combination of read conversion and genome conversion will yield successful alignment. For each UTR, both C to T and G to A converted alignments are expected to exist, and they are analyzed separately.
Wild-type and converted base counts are enumerated at each position in the library where the reference genome sequence encodes either C or G. Base identities not conforming to the expected wild-type or converted base at a position are not counted – these may be due to sequencing errors or polymorphisms.
Conversion biases in a single condition are identified by a) calculating a Z-score of WT base proportion at each informative position for an experimental condition (e.g., Late stage) (separate mean and standard deviation are used for C and G bases); and b) calculating a G test of independence for the number of WT and converted bases in the experimental condition versus control condition. A Benjamini-Hochberg control for false discovery rate is applied to the resulting p values. Positions with significant WT base depletion biases are defined to have Z < −1 and adjusted G-test p value < 0.05. For positions with significant WT base enrichment, a Z > 1 threshold is used.
Some bases due to their close proximity are effectively “linked” in their contribution to regulatory activity, e.g. the two Cs in the miR-430 seed (GCACUU) -- both GUACUU and GCAUUU should exhibit diminished destabilization efficacy, despite encoding one unconverted C. For linked bases, consecutive (but not necessarily adjacent) C or G positions are considered. Counts for fully wild-type (C-C or G-G for di-bases, C-C-C or G-G-G for tri-bases) versus mutated (at any one of the positions) alleles are enumerated across the library, then analyzed as above.
Meta-analyses for destabilization-enhancing base conversions were performed by using only the subset of sequencing reads that encode wild-type miR-430 motifs of interest, then counting base conversions at adjacent positions, where relevant (i.e., the reference sequence encodes a C or G base at that position). Base counts at the same relative positions were pooled across all instances of the motif in the library, and a G test of independence was performed for each position comparing late and early stages, for each of C to T and G to A conversions.
RESA-CLIP procedure
800 Zebrafish embryos were injected with 80pg RESA library and 80pg flag-tagged Ago2, crosslinked with 254 nm UV at 30% epiboly stage (3.3hfp) for 4 min on ice, and lysed in 750μL lysis buffer (50 mM Tris–HCl, pH 7.4, 100 mM NaCl, 1% Igepal CA-630, 0.5% sodium deoxycholate, 0.1% SDS, Protease Inhibitor (Roche # 04693159001) 1 tablet/10mL, 1:1000 volume of RNAse inhibitor (ThemoFisher #AM2696)), immunoprecipitated with 100μL Anti-Flag M2 Magnetic Beads (Sigma #M8823) for 2 hours at 4 °C, and washed 5 times with lysis buffer without protease inhibitor. To recover bound mRNAs beads were incubated with 200μL Proteinase K buffer (100mM Tris-HCl, pH 7.4, 50mM NaCl, 10mM EDTA) with 10μL Proteinase K (Thermo Fisher) for 20 min at 37 °C with shaking at 1100RPM. RNA was extracted with Trizol reagent. The entire RNA sample was reverse transcribed. Sequencing libraries were built as described above using 8 PCR cycles for input sample and 21 cycles for Flag IP.
Read alignment was performed as above for CLIP and input. To avoid PCR artifacts, each position along the UTRs was allowed to be overlapped by only 2 read fragment starts or ends, with the exception of the first and last position in each UTR, which had no restrictions.
Genetic deletion of 3′UTR regions
CRISPR-mediated mutagenesis was performed as described in30. Genotyping oligos used to amplify wild type and mutant alleles are (For AAAGCTGGAGGTTTGCAGAG, Rev CAAACTGAGGGACCTGGAGA) In situ hybridization was performed as in46 with 20ng of DIG-labeled RNA probe per 200μL hybridization reaction. RNA probes were synthesized from PCR product with T7 promoter on the reverse orientation oligo. For – CTGGGTGTCTCCTGGGTTTA, Rev – AGGCTGAGGGTGAAGCTGTA.
Reporter validation using qRT-PCR
All validation reporter constructs were generated using overlap-extension PCR by combining templates encoding SP6 promoter and GFP, SV40 polyadenylation signal, and tested insert with 15nt overhangs complementary to GFP on 5′ end and SV40 on 3′ end. RNA was in vitro synthesized using Sp6 Message Machine (ThermoFisher AM1340).
Zebrafish embryos were injected with 4pg of each reporter and dsRED control mRNAs and collected at 2hpf and 8hpf. Total RNA (250ng) was reverse transcribed using SuperScript III Kit (Invitrogen #18080-051) with random hexamers. cDNA was diluted 1:20 and used in 10uL reaction (5μL SYBR green master mix (ThermoFisher #4472908), 0.5μL 10μM Forward and Reverse primer mix, 1μL 1:20 diluted cDNA, 3.5μL water). All GFP reporters used a common forward primer CATGGTCCTGCTGGAGTTCGTGAC and a reporter-specific reverse primer (sdr16c5b CTAACTGTGTCTCTCTTCTTCACAG, shroom4 CCTCTCCCATCTGAACCAAC, rap2c CACTGCTCTTGCTTCTCGGCTCG, pam CGGTCGAAGCCATGTCG, itpr3 TTGAGGGTGTGTAGATCCAA). dsRED was amplified with Forward GAAGGGCGAGATCCACAAG and Reverse GGACTTGAACTCCACCAGGTA primers. Biological and technical triplicates were performed for each sample and relative expression with ΔΔCT method was measured using ViiA 7 Software v1.2.2 with dsRED mRNA as a reference control.
Zebrafish maintenance
Zebrafish embryos were obtained through natural mating of TU/AB strain of mixed ages (5–18 months). Mating pairs were selected randomly from a set of fish previously allocated to be used for the week(s) when embryos were collected. Number of mating pairs was determined by the total number of embryos required for analysis on that day, given approximately 100 embryos per mating pair. Embryos from multiple pairs are pooled prior to random allocation into experimental groups. No experimenter blinding was done. Fish were maintained in accordance with research guidelines of the International Association for Assessment and Accreditation of Laboratory Animal Care, under a protocol approved by the Yale University Institutional Animal Care and Use Committee (IACUC).
Supplementary Material
Acknowledgments
We thank K. Bilguvar, S. Mane, C. Castaldi and I. Tikhonova for sequencing support; H. Codore for technical help; A. Bazzini, P. Oikonomou and S. Tavazoie for discussions; all the members of the Giraldez laboratory for intellectual and technical support. This research was supported by the US National Institutes of Health grants (R01 HD074078, GM103789, GM102251, GM101108 and GM081602), Pew Scholars Program in the Biomedical Sciences, March of Dimes 1-FY12-230, the Yale Scholars Program, the HHMI Faculty Scholars Program, and the Whitman fellowship funds provided by E. E. Just, Lucy B. Lemann, Evelyn and Melvin Spiegel, The H. Keffer Hartline and Edward F. MacNichol, Jr. of the Marine Biological Laboratory in Woods Hole, MA to AJG; the Swiss National Science Foundation (grant P2GEP3_148600 to CEV); F32HD061194 CMT; F32HD071697 and start-up funds from the University of Pittsburgh to MTL; NIH Training grants T32 GM007223 and T32 HD007149, the Edward L. Tatum Fellowship (Yale University) and the Yale MRSP to VY.
Footnotes
Data Availability
Raw reads are publicly accessible in the Sequence Read Archive under SRP090954.
Code Availability.
Source code and executables for RESA are available at https://github.com/MTLeeLab/RESA
Author contributions:
VY, CMT and AJG designed and conceived the project. VY generated the RESA UTR, RESA-Bisulfite, and RESA-CLIP libraries, and the validation experiments. VY and MTL developed the RESA and RESA-Bisulfite analysis. MTL and CEV performed the computational analyses. All authors interpreted and analyzed the data. VY, MTL and AJG wrote the manuscript with input from the other authors.
Competing Financial Interests Statement:
The authors declare no competing financial interests.
References
- 1.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 2.Meola N, Gennarino VA, Banfi S. microRNAs and genetic diseases. Pathogenetics. 2009;2:7. doi: 10.1186/1755-8417-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giraldez AJ, et al. MicroRNAs Regulate Brain Morphogenesis in Zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 4.Lai EC, Posakony JW. The Bearded box, a novel 3′ UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development. 1997;124:4847–56. doi: 10.1242/dev.124.23.4847. [DOI] [PubMed] [Google Scholar]
- 5.Treisman R. Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5′ element and c-fos 3′ sequences. Cell. 1985;42:889–902. doi: 10.1016/0092-8674(85)90285-5. [DOI] [PubMed] [Google Scholar]
- 6.Pique M, Lopez JM, Foissac S, Guigo R, Mendez R. A Combinatorial Code for CPE-Mediated Translational Control. Cell. 2008;132:434–448. doi: 10.1016/j.cell.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 7.Mowry KL, Melton Da. Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science. 1992;255:991–4. doi: 10.1126/science.1546297. [DOI] [PubMed] [Google Scholar]
- 8.Zubiaga AM, Belasco JG, Greenberg ME. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giraldez AJ, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 10.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen CB, et al. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 13.Grimson A, et al. MicroRNA Targeting Specificity in Mammals: Determinants Beyond Seed Pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA – mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hafner M, et al. Transcritpome wide identification of RNA binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geisberg JV, Moqtaderi Z, Fan X, Ozsolak F, Struhl K. Global Analysis of mRNA Isoform Half-Lives Reveals Stabilizing and Destabilizing Elements in Yeast. Cell. 2014;156:812–824. doi: 10.1016/j.cell.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oikonomou P, Goodarzi H, Tavazoie S. Resource Systematic Identification of Regulatory Elements in Conserved 3′ UTRs of Human Transcripts. Cell Rep. 2014;7:281–292. doi: 10.1016/j.celrep.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenberg AB, Patwardhan RP, Shendure J, Seelig G. Learning the Sequence Determinants of Alternative Splicing from Millions of Random Sequences. Cell. 2015;163:698–711. doi: 10.1016/j.cell.2015.09.054. [DOI] [PubMed] [Google Scholar]
- 20.Weingarten-Gabbay S, et al. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science. 2016;354:240. doi: 10.1126/science.aad4939. [DOI] [PubMed] [Google Scholar]
- 21.Zhao W, et al. Massively parallel functional annotation of 3′ untranslated regions. Nat Biotechnol. 2014;32:387–91. doi: 10.1038/nbt.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wissink EM, Fogarty EA, Grimson A. High-throughput discovery of post-transcriptional cis-regulatory elements. BMC Genomics. 2016;17:177. doi: 10.1186/s12864-016-2479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolter JM, Kotagama K, Babb CS, Mangone M. Detection of miRNA Targets in High-throughput Using the 3′LIFE Assay. J Vis Exp. 2015:e52647. doi: 10.3791/52647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yartseva V, Giraldez AJ. Current Topics in Developmental Biology. Elsevier Inc; 2015. The Maternal-to-Zygotic Transition During Vertebrate Development: A Model for Reprogramming; p. 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee MT, et al. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013;503:360–364. doi: 10.1038/nature12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bazzini AA, Lee MT, Giraldez AJ. Ribosome Profiling Shows That miR-430 Reduces Translation Before Causing mRNA Decay in Zebrafish. Science. 2012;336:223–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staton AA, Knaut H, Giraldez AJ. Reply to: ‘On the robustness of germ cell migration and microRNA-mediated regulation of chemokine signaling’. Nat Genet. 2013;45:1266–1267. doi: 10.1038/ng.2812. [DOI] [PubMed] [Google Scholar]
- 28.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:0404–0418. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vella MC, Choi E, Lin S, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno-Mateos MA, et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment:RNA ligands to bacteriophage T4 DNA polymerase. Science (80- ) 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 32.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–22. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 33.Blackwell TK, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 34.Martin KC, Ephrussi A. Review mRNA Localization: Gene Expression in the Spatial Dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabani M, Kertesz M, Segal E. Computational prediction of RNA structural motifs involved in post-transcriptional regulatory processes. Methods Mol Biol. 2011;714:467–479. doi: 10.1007/978-1-61779-005-8_28. [DOI] [PubMed] [Google Scholar]
- 36.Bazzini AA, et al. Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J. 2016:e201694699. doi: 10.15252/embj.201694699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mishima Y, Tomari Y. Codon Usage and 3′ UTR Length Determine Maternal mRNA Stability in Zebrafish. Mol Cell. 2016;61:874–885. doi: 10.1016/j.molcel.2016.02.027. [DOI] [PubMed] [Google Scholar]
- 38.Presnyak V, et al. Codon optimality is a major determinant of mRNA stability. Cell. 2015;160:1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis CA, Monnier JM, Nick HS. A Coding Region Determinant of Instability Regulates Levels of Manganese Superoxide Dismutase mRNA *. J Biol Chem. 2001;276:37317–37326. doi: 10.1074/jbc.M104378200. [DOI] [PubMed] [Google Scholar]
- 40.Wellington CL, Greenberg ME, Belasco JG. The Destabilizing Elements in the Coding Region of c-fos mRNA Are Recognized as RNA. Mol Cell Biol. 1993;13:5034–5042. doi: 10.1128/mcb.13.8.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozen S, Skaletsky H. Primer3 on the WWW for General Users and for Biologist Programmers. Bioinforma Methods Protoc Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 42.Jiang H, Lei R, Ding S, Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:1–12. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bailey TL, et al. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.