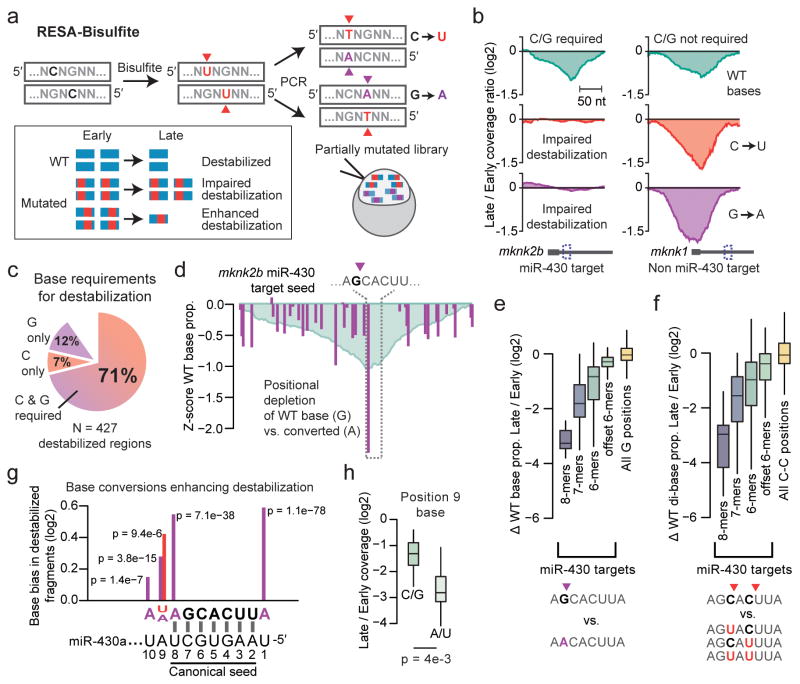
Figure 3. RESA-Bisulfite implements high-throughput mutational analysis for regulatory sequences.
(a) RESA-Bisulfite used bisulfite treatment to induce C to U or G to A mutations within the reporter library pool. Partially mutated reporters are injected in vivo to determine the effect of mutations (red, purple) on the destabilization of wild type (WT, blue) regions. (b) Coverage ratio plots for reads with wild-type bases (top), C to U converted bases (middle), and G to A converted bases (bottom) for a region within mknk2b that requires C and G bases for destabilization (left) and a region within mknk1 that does not require C or G bases for destabilization (right). (c) Proportion of destabilized regions identified by RESA that require C and/or G bases for destabilization. (d) Positional analysis of G to A conversions over the mknk2b miR-430 target seed at late stage, showing positions with G depletion (purple). Y-axis measures the Z score of the ratio of WT base count to mutated base count at each position, compared to all positions. RESA signal is overlaid in blue. (e,f) Depletion of wild-type G bases versus converted A (e) and C-C dibases versus converted U-C, C-U, and U-U (f) in late-stage versus early-stage embryos across all miR-430 targets grouped by seed complementarity: 8-mers (N=13), 7-mers (N=40), 6-mers (N=136), offset 6-mers (N=115). All G (N=99157) and C-C (N=92696) positions across the library also shown. Boxes show median and quartiles, whiskers show 1.5 times the interquartile range. Outliers not shown. (g) Pooled base conversion biases adjacent to miR-430 target seeds showing enrichment of wild-type bases relative to converted. Y-axis shows the magnitude of change in the proportion of converted bases observed (U, red; A, purple) from early to late stage. X-axis is oriented sense to the target. P value from a G test of independence is shown. Axes as in (h). (j) Boxplots comparing RESA destabilization of miR-430 targets encoding endogenous A or U at position 9 (N=20) versus C or G (N=22) for 8-mers and 7-mers (pairing positions 2–8). Boxes show median and quartiles, whiskers show 1.5 times the interquartile range. Outliers not shown.