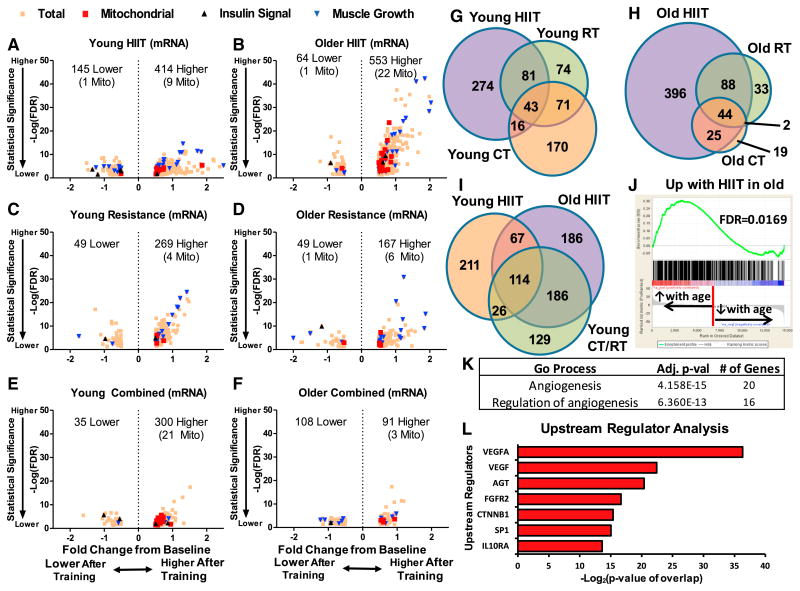
Figure 4. Muscle Gene Expression Changes with Exercise Training.
(A–F) Genes that were differentially expressed following high-intensity interval training (HIIT) in the young (A) or older (B), resistance training in the young (C) or older (D), and combined training in the young (E) or older (F) using an adjusted p value of ≤0.05 and an absolute fold change of ≥0.5 were annotated according to their mitochondrial specificity (using MitoCarta) and molecular function (using KEGG). Mito stands for mitochondrial.
(G and H) Overlap of genes upregulated with different modes of exercise training in younger (G) and older (H) participants.
(I) Overlap of genes that were upregulated with HIIT in older adults and any type of exercise training in younger adults.
(J) Gene set enrichment analysis of baseline gene expression differences between young and old participants against genes that were upregulated with HIIT in older participants. Genes that increased expression with age were more likely to increase their expression with HIIT in older participants.
(K) A “universal exercise training response gene set” was derived by looking for genes that increased with exercise with an adjusted p value of ≤0.05 and an absolute fold change of ≥0.3 in all groups. Gene Ontology (GO) process annotations enriched for this universal exercise training response gene set was derived using MetaCore software configured with an adjusted p value threshold of ≤0.05.
(L) Ingenuity Pathway Analysis (QIAGEN) was used to detect up stream regulators of the “universal exercise training response gene set.”