Figure 6. Downregulation of mRNA while Protein Abundance Increased for Mitochondrial and Ribosomal Protein with HIIT in Older Adults.
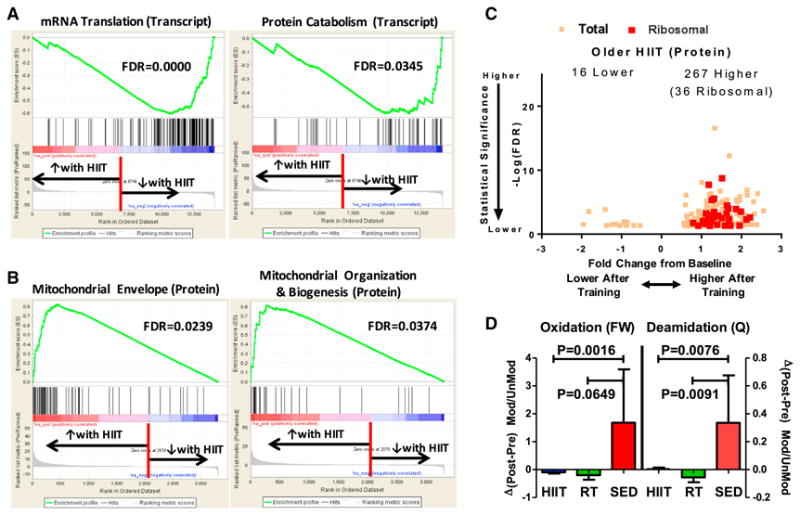
(A) Gene set enrichment analysis of transcripts reveal downregulation ofmRNA for translational and protein catabolism pathways between pre-HIIT and post-HIIT in older cohort (adjusted p value < 0.05 and an absolute log2 fold change ≥ 0.5).
(B) Gene set enrichment analysis of proteins demonstrate an increased abundance of mitochondrial proteins between pre-HIIT and post-HIIT in older cohort (adjusted p value < 0.05 and an absolute log2 fold change ≥ 0.5). The software was configured to find enrichment from predefined gene sets in MSigDB.
(C) Ribosomal protein abundance increased with HIIT in older, supporting a role for greater translational capacity. MaxQuant software configured to process label-free data from older HIIT group was used to detect differentially expressed proteins with an adjusted p value of ≤0.05 andan absolute fold change of ≥0.5. Mitochondrial ribosomal proteins are highlighted in red and detailed in Table S7.
(D) Post-translational modification analysis of older subjects showed decreased oxidative damage to proteins after exercise training when compared to sedentary (SED) state. For this, MS1 intensities of peptides with phenylalanine oxidation, tryptophan oxidation, and glutamine deamidation were used to compute the ratio of modified (Mod) to unmodified (UnMod) quantities. Distribution of ratio of Mod/UnMod for all peptides was plotted and (two-sided) p values were computed using Mann-Whitney rank-sum test.
