Figure 1.
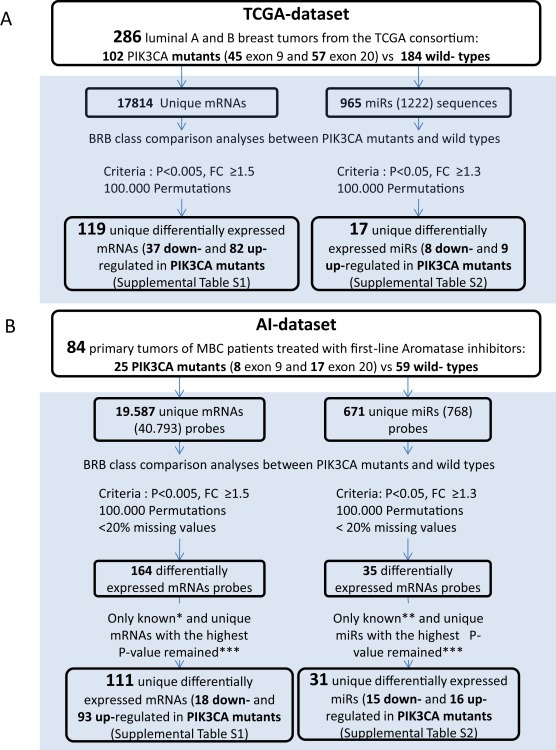
Workflow used to discover differentially expressed mRNAs and miRs when analyzed by PIK3CA mutation status. A) TCGA dataset (publicly available data from the TCGA consortium). B) AI dataset. *Known mRNAs indicates mRNAs with gene names annotated according to platform GPL6480 updated in Nov 16, 2014 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL6480); **known miRs indicates miRs annotated according to miRbaseV20; ***unique mRNAs or miRs with the highest P‐value indicate those cases in which at least two probes for an annotated mRNA or miR name were found differentially expressed. We selected the probe with the highest P‐value.
