Abstract
1. Purpose
We investigated whether microRNA expression data from glioblastoma could be used to produce a profile that defines a bevacizumab responsive group of patients.
2. Patients and methods
TCGA microRNA expression data from tumors resected at first diagnosis of glioblastoma in patients treated with bevacizumab at any time during the course of their disease were randomly separated into training (n = 50) and test (n = 37) groups for model generation. MicroRNA‐seq data for 51 patients whose treatment included bevacizumab in the BELOB trial were used as an independent validation cohort.
3. Results
Using penalized regression we identified 8 microRNAs as potential predictors of overall survival in the training set. We dichotomized the response score based on the most prognostic minimum of a density plot of the response scores (log‐rank HR = 0.16, p = 1.2e−5) and validated the profile in the test cohort (one‐sided log‐rank HR = 0.34, p = 0.026). Analysis of the profile using all samples in the TCGA glioblastoma dataset, regardless of treatment received, (n = 473) showed that the prediction of patient benefit was not significant (HR = 0.84, p = 0.083) suggesting the profile is specific to bevacizumab. Further independent validation of our microRNA profile in RNA‐seq data from patients treated with bevacizumab (alone or in combination with CCNU) at glioblastoma recurrence in the BELOB trial confirmed that our microRNA profile predicted patient benefit from bevacizumab (HR = 0.59, p = 0.043).
4. Conclusion
We have identified and validated an 8‐microRNA profile that predicts overall survival in patients with glioblastoma treated with bevacizumab. This may be useful for identifying patients who are likely to benefit from this agent.
Keywords: microRNA, Glioblastoma, Bevacizumab, Glioma, Prediction
Highlights
An 8‐microRNA algorithm predicts glioblastoma response to bevacizumab.
The predictive value was bevacizumab specific.
The algorithm was independently validated using BELOB trial patients.
1. Introduction
Bevacizumab (BEV) is an anti‐angiogenic monoclonal antibody that acts by slowing the growth of new blood vessels in tumors through inhibition of VEGFA (vascular endothelial growth factor A) (Glade‐Bender et al., 2003). In glioblastoma, two prospective, randomized, placebo controlled clinical trials, AVAglio and RTOG 0825, have been performed to assess whether BEV improves survival in patients with newly diagnosed glioblastoma (Chinot et al., 2014; Gilbert et al., 2014). Both studies reported improved progression‐free survival (PFS) but no overall survival (OS) benefit. Despite these results there is anecdotal evidence, and some evidence from observation of tails of Kaplan Meier survival curves, that certain patients may benefit from BEV treatment. Identification of these patients is an unmet need (Field et al., 2014; Prados et al., 2015).
Prediction of benefit from BEV in glioblastoma patients has been attempted previously. Colman et al. developed a prognostic signature for glioblastoma (all treatments) which was based on expression of genes associated with mesenchymal differentiation and angiogenesis (Colman et al., 2010). This was then assessed using tumor samples from the RTOG 0825 patients3. The results did not show the expected association between worse OS and PFS with the mesenchymal subtype.
A smaller phase II trial (the BELOB trial) in the Netherlands assessed BEV or CCNU mono‐or combination therapy in recurrent glioblastoma, with a primary endpoint of OS (Taal et al., 2014). However, survival benefit in glioblastoma could not be confirmed in the phase III EORTC 26101 trial (Wick et al., 2015). In this trial, patients with progressive disease after standard chemo‐radiotherapy with temozolomide were randomized 2:1 between CCNU 90 mg/m2 mg every six weeks plus 10 mg/kg bevacizumab every two weeks and CCNU single agent 110 mg/m2 every six weeks followed by investigators choice at further progression. Although the progression free survival was improved in the combination arm, there was no overall survival benefit.
When patients from the BELOB trial were assigned to molecular subtypes results showed that the EGFR amplified, classical glioblastoma subtype responded well to the combination therapy and the mesenchymal subtype showed a poor response to combination therapy. It should be noted that these data included only 28 patients in the mesenchymal single agent BEV group, and they are not yet fully published (Eraslan et al., 2014). Overall, these, and other data may suggest that anti‐angiogenic therapy resistance is associated with the mesenchymal transition, and that tumors with more infiltrative phenotypes are more resistant to these drugs (Piao et al., 2013, 2012). Retrospective analysis of the AVAglio trial showed patients with IDH wild‐type proneural tumors had improved OS when treated with BEV first‐line, and these are the most encouraging data linking tumor sub‐type to outcome thus far (Omuro et al., 2014).
Further analysis of translational data from the AVAglio trial suggested that neither VEGFA or VEGFR2 (vascular endothelial growth factor receptor 2) are predictive or prognostic biomarkers in the context of BEV treatment, although a VEGFA SNP rs2010963 is associated with vascular toxicity (Di Stefano et al., 2014; Field et al., 2014). Plasma levels of MMP2 (matrix metalloproteinase 2) have been shown to be associated with response and survival in BEV‐treated patients (single agent therapy) in a study by the Chinot lab (Tabouret et al., 2014).
MicroRNAs have not been studied as predictive indicators for BEV response to date. Their stability in clinical samples and role in glioma biology suggest they represent prime candidates for use in predictive signatures/profiles (Hall et al., 2012; Hayes et al., 2014).
In this study, we have attempted to identify a prognostic microRNA profile in BEV treated patients using OS as an endpoint. Our results show that an 8‐microRNA profile can define patients treated with BEV who have a better prognosis.
2. Materials and methods
2.1. TCGA clinical information and expression data
Level 3 Agilent microRNA 8 × 15 k microarray expression data plus clinical and treatment information for 563 glioblastoma samples, 90 of which were from patients treated with BEV used either as an adjuvant with first‐line treatment, at progression or recurrence, were downloaded from TCGA (Cancer Genome Atlas Research Network, 2008). Patients had been treated using varying numbers of 2–3 week cycles of BEV therefore treatment time was determined as the date from the start of treatment to the date of the end of treatment. Samples were taken at diagnosis and OS was measured from the date of diagnosis, regardless of timing of BEV treatment (however timing of treatment was analyzed as a variable in a separate multivariable analysis). Three patients were removed due to lack of start date information, resulting in a total of 87 patients. These were randomly split into test and training set groups of 50 and 37 patients respectively (Table 1). These numbers were chosen to maximize power in generation of the model, whilst allowing a sufficient validation cohort for testing of the model.
Table 1.
Summary of data from the test and training set cohorts. Samples from TCGA were randomly split into training and test sets of 50 and 37 patients respectively. The test set has a marginally poorer prognosis and KPS and, on average, 28.5 days shorter treatment time for BEV. Additionally, 22% of test set patients were treated with BEV as an adjuvant treatment, whereas only 16% of patients in the training set were treated as an adjuvant treatment. Days to death are recorded where possible, and where the patient was living at the end of the data collection, days to last follow‐up were used.
| Training set (n = 50) | Test set (n = 37) | |
|---|---|---|
| Age | Median 54.5 years | Median 56 years |
| <60 years | 33 | 27 |
| ≥60 years | 17 | 10 |
| Gender | ||
| Male | 27 | 23 |
| Female | 23 | 14 |
| Karnofsky performance score | ||
| ≤70 | 21 | 18 |
| >70 | 29 | 19 |
| Days to death/last follow‐up | ||
| <450 days | 25 | 23 |
| ≥450 days | 25 | 14 |
| <30 days | 0 | 0 |
| Treatment regimen | ||
| Adjuvant | 8 | 8 |
| Progression | 29 | 16 |
| Recurrence | 5 | 2 |
| Not available | 8 | 11 |
| Mean treatment length | 205.9 | 177.4 |
2.2. Generation of a risk algorithm for OS in bevacizumab‐treated glioblastoma patients using microRNAs
The training set samples were assessed using LASSO penalized regression (Tibshirani, 1996) with leave‐one‐out cross‐validation using R software (v2.15.1) and the Penalized package (Goeman, 2010). This produced 8 microRNAs with non‐zero coefficients.
A response score was generated using the sum of microRNA expression values weighted by the coefficients from the LASSO regression.
This was: E_miR‐n = expression of microRNA n.
The response score was applied to all samples in the training set. The most prognostic cut‐off was chosen based on log‐rank tests at each minimum value on a density plot. The rationale behind this is that biologically it is assumed that patients either show some benefit to the treatment or not. The training set samples were then separated into responders and non‐responders using this cut‐off. A Cox regression model incorporating age and the log‐rank test were used to assess OS of the two groups in the training set. 200 permutations of a 50‐patient training set from the 87 original patients were used as input to LASSO to determine model differences with different patients. The response score was also assessed as a predictor of PFS. A statistical significance threshold of p = 0.05 was used throughout, with two‐tailed log rank tests for the training set and one‐tailed tests for all validations. The length of treatment time was tested for correlation with the survival time in both responder and non‐responder groups. Fisher's exact test was also performed on the responder groups for the molecular subtype, treatment regimen and histological features.
2.3. Validation of the response score in the test set
The response score was calculated with the above algorithm using the microRNA expression values for the 37 test set samples. The defined cut‐off from the training set of a response score of 0 was used to separate the test set into two groups of responders and non‐responders. A Cox regression model incorporating age and the log‐rank test were used to assess OS of the responder groups. The length of treatment time was assessed for correlation with survival time in both responder and non‐responder groups. Multivariate analysis of other prognostic indicators assessed in the trial was performed to determine whether the responder groups are independent prognosis predictors.
2.4. Testing of the algorithm across all treatment types
The response score was applied to all 473 patients in the TCGA (treated with various treatment regimens not including BEV) (Table S1). This cohort was split into two responder groups based on the response score cut‐off of 0 and the two groups were assessed by Cox regression and log‐rank test.
2.5. Validation of the profile using BELOB trial data
Patients were eligible for the BELOB trial if they were ≥18 years and had a first recurrence of glioblastoma after temozolomide and radiotherapy treatment. Details of the study have been described previously (Taal et al., 2014). Total RNA extraction, purification, and quantification from formalin‐fixed and formalin‐fixed paraffin‐embedded (FFPE) material were reported previously (Gravendeel et al., 2011). 500 ng RNA was used for sequencing on an Illumina TruSeq and ∼35–40 million 40 base paired end‐reads were generated per sample. RNA‐seq (n = 96) was run by Expression Analysis (Durham, NC). Gene expression levels (Ref‐seq genes) were extracted from the RNA‐seq data using featureCounts (Liao et al., 2014), after alignment on hg19 with Tophat2 (Trapnell et al., 2009) of clipped/trimmed reads as provided by the manufacturer. The response score was calculated using read per million counts and the cut‐off value was defined by using the minimum value of a density plot of the response scores.
2.6. Pathway analysis of the microRNAs
DIANA microT (Paraskevopoulou et al., 2013) target predictions for the microRNAs and the DIANA miRPath pathway analysis tool (Vlachos et al., 2012) were used to identify the pathways enriched for predicted targets of the eight microRNAs in the signature. The union of the predicted targets was used for the pathway enrichment and p‐values were corrected for multiple tests using Benjamini‐Hochberg's FDR.
3. Results
3.1. An 8‐microRNA profile generated from the training set predicts prognosis in bevacizumab treated patients
Using the LASSO method, 8 microRNAs were identified with non‐zero regression coefficients in our training dataset of 50 glioblastomas. A response score was created using the algorithm stated in methods. The response score was then plotted as a density plot (Figure 1). The response score itself showed a normal distribution (Figure S1) and therefore the minimum values of a density plot at a bandwidth of 0.009 were used as a guide for determining a cut‐off. The response score value at each minimum of the density plot was determined and a log‐rank test was performed with this value as a cut‐off to determine an optimal cut‐off for dichotomization. The minimum density that occurred around a response score of 0 showed the highest significance with the best hazard ratio, and so a response score of 0 was chosen for the cut‐off (Figure 2A, Table S2). This cut‐off is justified because the expression data are quantile normalized, and therefore a cut‐off of 0 represents the expression of each of the microRNAs in the signature at median survival of all patients. The median survival time of the responder group defined in this way was 22 months and the median of the non‐responder group was 12 months. A log rank test performed on 200 permutations of the 50‐patient training set was significant for predicting altered survival in 100% of tests with p < 0.05 and in 96.5% of tests with p < 0.005.
Figure 1.
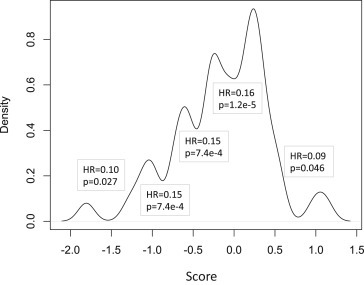
Density plot of the response scores to determine a cut‐off for dichotomization. The response score, calculated according to the microRNA profile algorithm was illustrated as a density plot (bandwidth 0.009). This shows the fraction of patients with scores at each value on the X‐axis and was used to determine whether a natural cut‐off of the response score could be ascertained for dichotomization of the score into two groups of responders and non‐responders. As multiple minima were identified from the density plot, the optimal minimum was determined by assessing prognostic ability. Each minimum value was used as a cut‐off to define two ‘response’ groups. These two groups were then assessed for differences in survival using the log rank test. The hazard ratio and p‐value at each minimum are shown on the plot. The most significant association with survival occurred when a cut‐off score of 0 was used which corresponds to the baseline hazard determined by the microRNAs.
Figure 2.
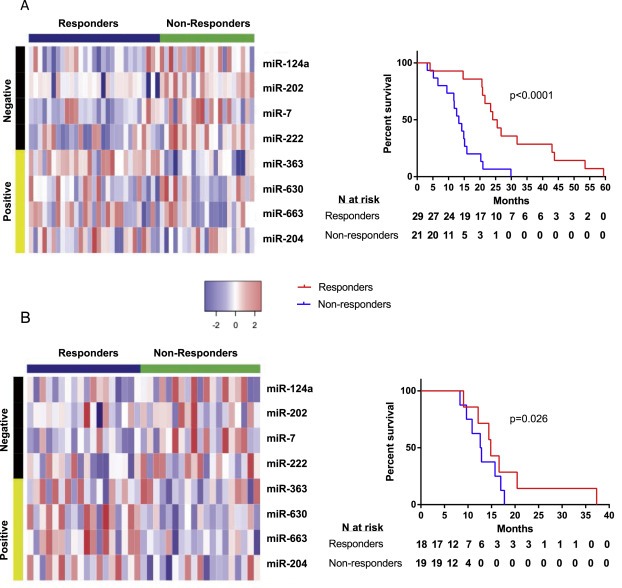
MicroRNA expression and patient survival in the responder and non‐responder groups in the test and training sets. The responder and non‐responder groups were calculated using the microRNA profile and split into two responder groups using a cut‐off score of 0. (A) A heat map and survival curve of microRNA expression of the 8 profile microRNAs and survival of the responder and non‐responder groups in the training set. Negative microRNAs are those that are higher in samples from patients with poorer survival and conversely, positive microRNAs are those that are lower in patients with poorer survival. The accompanying survival curve confirms that patients who stratified to the responder group using the profile had a better outcome than those stratified to the non‐responder group. (B) A heat map and survival curve of microRNA expression of the 8 profile microRNAs and survival of the responder and non‐responder groups in the test set. A one‐tailed log‐rank test showed that patients stratified to the responder group in the test set had a better outcome than those in the non‐responder group, as shown in the accompanying survival curve.
Spearman's correlation of duration of BEV treatment with survival time showed that the responders showed a correlation (correlation coefficient = 0.48, p = 0.01) whereas the non‐responders did not (correlation coefficient = 0.36, p = 0.12). Multivariable Cox regression of the responder group and age showed the responder group to be an independent predictor of survival irrespective of age (group HR = 0.11, CI = 0.04–0.29, p = 5.4e−6, age HR = 1.03, CI = 1.00–1.06 p = 3.3e−2).
3.2. Assessment of the profile in the test group of 37 patients
Response scores were calculated for the 37 test set patients using a cut‐off of 0 to separate the patients into responder and non‐responder groups. This produced a group of 18 responders, with a median survival 21 months and a group of 19 non‐responders with a median survival of 15 months. A one‐sided log rank test showed that the responders survived significantly longer than the non‐responders (HR = 0.34, CI = 0.11–1.01, p = 0.026, Figure 2B). Multivariable Cox regression with age confirmed that the responder group was a prognostic factor (HR = 0.33, CI = 0.11–0.99, p = 0.049) independent of patient age.
3.3. Testing of the profile across all the glioblastoma patients in the TCGA database, independent of treatment
To test whether our profile is predictive of patient outcome in general or specific to BEV, we calculated the response score for all 473 glioblastoma patients in the TCGA database. These were treated with various drugs and regimes not including those treated with BEV (Table S1). This identified 256 patients in the responder group (median survival 9.25 months) and 217 in the non‐responder group (median survival 7.55 months). A two‐sided log rank test between the responder groups showed this profile is not prognostic for OS (HR = 0.84, CI = 0.68–1.02, p = 0.083, Figure 3). This indicates the profile is predicting benefit from BEV specifically.
Figure 3.
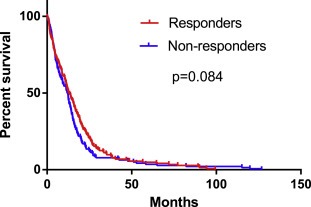
The survival of the profile‐defined responder and non‐responder groups when calculated for all glioblastoma patients in the TCGA. In order to determine whether the profile‐based definitions of responder and non‐responder groups are specific to patients treated with BEV, the response score was calculated and a cut‐off score of 0 was used to test all the patients in the TCGA, regardless of their treatment (not including those patients treated with BEV). The profile‐defined responder groups were not associated with prognosis (HR BEV‐treated training set = 0.16, HR bev‐treated test set 0.34, HR all treatments = 0.84). This suggests that the profile predicts prognosis more strongly in BEV treated glioblastoma.
3.4. Characterization of the responder groups defined by the profile
We determined the proportions of each molecular glioblastoma subtype in the responder and non‐responder groups of the combined test and training sets. Analysis using Fisher's exact test showed that there were significantly fewer mesenchymal type tumors in the responder group (p = 0.041). The other subtypes did not show any significant difference between the responder and non‐responder groups (classical glioblastoma p‐value = 0.15, neural glioblastoma p‐value = 0.56, proneural G‐CIMP glioblastoma p‐value = 1.00, proneural non‐G‐CIMP glioblastoma p = 1.00). Multivariable Cox regression using the microRNA profile and subtype (as the only variables in the model) showed that the microRNA profile was independent of subtype using a two‐tailed test in the training set (HR = 0.13, 95% CI = 0.05–0.37, p = 1.1e−4) and using a one‐tailed test in the test set (HR = 0.28, 95% CI = 0.07–1.15, p = 0.04). Previous data have suggested that molecular subtype may be predictive of benefit from BEV (Eraslan et al., 2014); however, the microRNA‐based BEV response profile we have identified here has more predictive power than the molecular glioblastoma subtypes when directly tested.
The cohort of BEV treated patients includes different treatment start points during the course of a patient's disease. There was no significant difference in any treatment regime between the responder groups when tested with Fisher's exact test (adjuvant p‐value = 0.79, progression p‐value = 1.00, recurrence p‐value = 0.45). Multivariable Cox regression using the microRNA profile and subtype showed that the microRNA profile was independent of treatment time using a two‐tailed test in the training set (HR = 0.12, 95% CI = 0.04–0.32, p = 2.4e−5). A similar trend was observed in the test set (HR = 0.44, 95% CI = 0.14–1.31, p = 0.07) (although treatment time data were available for only 70% of patients in the test set cohort).
3.5. Ability of the profile to predict progression free survival
We tested whether the 8‐microRNA profile predicts PFS using Cox regression in both the test and training sets. As the cut‐off for PFS may be different than that for OS, the score was also assessed for ability to predict survival. The training set showed that decreasing response score predicts PFS (HR = 0.37, 95% CI = 1.42–5.03, p = 0.0024) and the dichotomized responder groups have significantly different PFS by log‐rank test (HR = 0.48, 95% CI = 0.25–0.93, p = 0.029). In the test set, decreasing response score predicted PFS (HR = 0.44, 95% CI = 1.02–5.04, p = 0.045) but the dichotomized responder groups did not (HR = 0.58, 95% CI = 0.29–1.13, p = 0.11). This may be because of the small size of the test set.
3.6. Validation of the profile using data from the BELOB trial
To independently validate the profile we calculated response scores for each of the patients in the BELOB trial using the microRNA algorithm. This was done using aligned reads from RNA‐seq data and was performed blind, with no knowledge of clinical data. An identical cut‐off to that in the training set was not possible, because this used microRNA reads from an RNA‐seq dataset and could not be normalized in the same way as the microarray data from the TCGA. The response score was plotted on a density plot and the minimum value of this density plot used as a cut‐off to generate two response groups (Figure 4A–B). One‐tailed log‐rank tests in all arms of the trial, and in just the arms that included bevacizumab as a treatment (either monotherapy or in combination with CCNU) are shown in Figure 4 and patient OS data from the BELOB trial are shown in Figure S2. In the BEV treated arms, the responder groups were significantly associated with survival (HR 0.59, 95% CI 0.32–1.09, p = 0.043) validating that the profile delineates patient groups with differing benefit from BEV. Log rank test of the predicted responders from the BEV arms with the predicted non‐responders from the CCNU showed no significance (HR 1.12, 95% CI 0.51–2.49, p = 0.36, Figure S3), which may suggest that the profile specifically defines a group of patients who do not benefit when treated with BEV. These results indicate that even the patient responder group treated with BEV show no more improvement in survival than those from the non‐responder group treated with CCNU.
Figure 4.
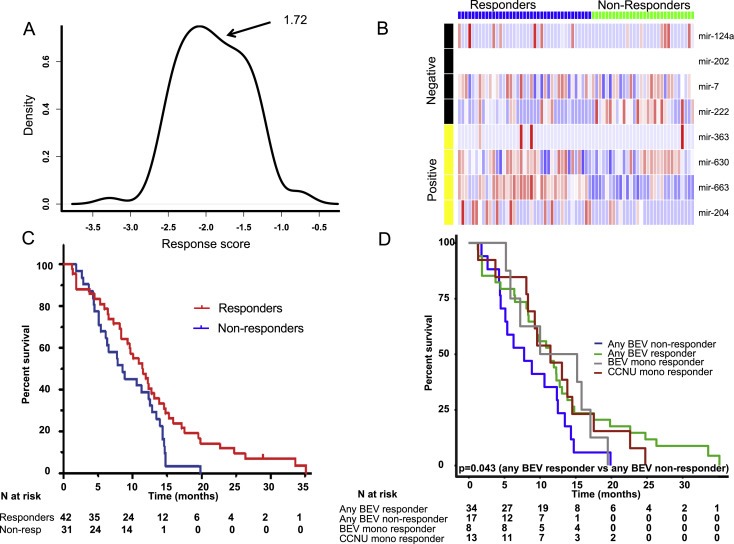
Survival curves for the profile‐defined responder and non‐responder groups in different arms of the BELOB trial. In order to validate the profile, the microRNA profile response score was calculated for patients in the BELOB trial who were treated with BEV (either as monotherapy or in combination with CCNU). (A) A density plot at bandwidth 0.009 of the response scores showed one minimum at a response score of 1.71 and this was used as a cut‐off to dichotomize. The calculation was performed blind, with the investigator having no knowledge of the clinical details of the patients. (B) MicroRNA expression of the response groups in the validation group. Negative microRNAs are those that are higher in samples from patients with poorer survival and conversely, positive microRNAs are those that are lower in patients with poorer survival. MiR‐202 sequences were not detected in this group but microRNA reads are of low abundance in RNA‐seq data and miR‐202 may therefore be expressed in these samples. (C) Survival of responder and non‐responder groups in all treatment arms from the BELOB trial (n = 73). (D) Survival of responder and non‐responder groups in the arms that included BEV as a treatment (monotherapy or in combination with CCNU, n = 51). The responder groups from other arms in the trial have been included for clarity.
Clinico‐pathological markers including age, gender, MGMT methylation status, IDH1 mutation status, were assessed by univariate and multivariate analysis in the whole trial and this showed that the responder groups defined by the profile were independently associated with survival (Table 2).
Table 2.
Univariate and multivariate analysis of parameters from the validation group. Using univariate analysis, the microRNA profile, Karnofsky performance score (KPS), MGMT methylation (MGMT) were also associated with survival in the trial cohort. Four different multivariate analyses, using survival and one other variable stated in the table, showed that the microRNA profile and KPS were independently associated with survival (although only KPS was significant after multiple testing correction (p = 0.042)). IDH1 refers to IDH1 mutation status.
| Variable | Number of subjects | HR | 95% CI | p‐value |
|---|---|---|---|---|
| Univariate analysis | ||||
| MGMT | 71 | 0.59 | 0.36–0.96 | 0.034 |
| IDH1 | 70 | 0.55 | 0.23–1.29 | 0.169 |
| KPS | 73 | 1.82 | 1.17–2.82 | 0.007 |
| Age | 73 | 1.01 | 0.99–1.03 | 0.546 |
| Gender | 73 | 0.87 | 0.54–1.41 | 0.579 |
| miR profile | 73 | 0.58 | 0.35–0.95 | 0.031 |
| Variable | HR | 95% CI | p‐value | |
| Multivariate analysis (68 subjects) | ||||
| MGMT | 0.71 | 0.43–1.18 | 0.185 | |
| KPS | 2.02 | 1.31–3.12 | 0.001 | |
| IDH | 0.74 | 0.31–1.77 | 0.493 | |
| miR profile | 0.59 | 0.35–0.99 | 0.046 | |
3.7. Pathway analysis of the microRNAs in the signature
DIANA microT (Paraskevopoulou et al., 2013) target predictions for the microRNAs and the DIANA miRPath pathway analysis tool (Vlachos et al., 2012) were used to determine which pathways the predicted targets of the microRNAs are enriched in. This showed that the mTOR and neurotrophin signaling pathways are enriched for targets of these microRNAs (Table S4). Neurotrophin signaling factors such as brain‐derived neurotrophic factor (BDNF) stimulate VEGF production and this effect is blocked using mTOR inhibitors (Nakamura et al., 2006). Interestingly, one of the microRNAs in the signature, miR‐204, is suppressed by BDNF, resulting in actin reorganization through the mTOR pathway (Imam et al., 2012).
4. Discussion
This study has identified an 8‐microRNA profile that has the potential to improve selection of patients for BEV treatment. This profile is preferable to previously published signatures/profiles because it uses only 8 predictors, which can easily be assayed in a clinical setting, using stable genetic markers (microRNAs) and has better prediction power than other factors such as MGMT promoter methylation status, IDH mutation and molecular subtype.
In order to further validate this approach for clinical application, it will be necessary to demonstrate that it is practical in a clinical context and in an appropriate time frame to permit treatment decisions to be made. Other investigators have shown that microRNA signatures can be used as predictive assays (Bucay et al., 2015; Rice et al., 2015). We would therefore suggest that this signature could be applied to glioma patients, for example using tumor RNA‐seq data, which is becoming increasingly available in a clinical context. By normalizing individual data to control samples, patients could then be categorized into likely responders versus non‐responders. The ultimate validation of this assay in predicting individual treatment responses requires assessment in prospective clinical studies.
Comparing our data with other reports suggests some important differences. Sandmann et al. reported that proneural IDH‐ wild‐type tumors might respond to first line BEV treatment (Sandmann et al., 2015). We did not find that there were more proneural non‐G‐CIMP patients in the responder group. This may be because this group is not highly represented in our data. There were fewer patients with tumors of mesenchymal molecular glioblastoma subtype in the responder category although 38% of mesenchymal tumors still stratified to the responder group, which indicates that the profile is not simply predicting a mesenchymal subtype.
If the microRNAs in the profile are associated with BEV response it is assumed they have a role in angiogenesis, and this was shown by collective analysis of the predicted targets. The microRNAs that have a positive weight in the profile (miR‐7, miR‐124a, miR‐202 and miR‐222), and therefore are lower in responders, are likely to be anti‐angiogenic. This is because responders should have more angiogenic tumors than non‐responders. The converse is also likely to be true for the negatively weighted microRNAs (miR‐204, miR‐663, miR‐630 and miR‐363).
Consistent with this hypothesis, of the eight microRNAs identified in the profile, seven are reported in the literature to be involved in angiogenesis. Of the positively‐weighted microRNAs, miR‐124a has been shown to transcriptionally decrease VEGF through RAS signaling (Shi et al., 2014) and miR‐222 is considered one of three most important anti‐angiogenic microRNAs in coronary artery disease (Zhang et al., 2011). Overexpression of miR‐7 in a neuroblastoma mouse model significantly reduced angiogenesis and in endothelial cell lines miR‐7 overexpression decreased tube formation and sprouting (Babae et al., 2014). Of the microRNAs that were negatively weighted, miR‐363 and miR‐663 are reported to improve angiogenesis and endothelial interaction with hematopoietic precursors (Costa et al., 2013) and miR‐663 indirectly increases VEGF and promotes angiogenesis (Afonyushkin et al., 2012). The reports on these microRNAs are concordant with their effect in the profile. However, miR‐204 and miR‐630 show anti‐angiogenic properties and are negatively weighted microRNAs; miR‐204 directly decreases VEGF and also targets angiopoietin‐1 (Kather et al., 2014; Zhao et al., 2014), and miR‐630 has been shown to be induced by the anti‐angiogenic protein angiopoietin‐like protein 1 (Kuo et al., 2013). These unexplained functions of certain microRNAs in the signature are likely reflective of the complex biology involved in the response of a patient and their tumor to BEV.
In addition to these associations with angiogenesis, miR‐202 is predicted to target FMO4 (Lewis et al., 2003), a drug metabolism gene associated with BEV response (Erdem‐Eraslan et al., 2016). This suggests that the effects these microRNAs have in patient response to BEV extend further than angiogenesis.
In summary, we have defined a promising approach to predicting response to BEV in GBM patients and further studies are warranted to test this profile further, in larger cohorts using clinically relevant assays.
Financial support
Josie Hayes was a Yorkshire Cancer Research (UK Registered Charity#516898) funded PhD student.
Conflict of interest
The authors have no conflicts of interest to declare.
Supporting information
The following are the supplementary data related to this article:
Figure S1. A) The distribution of the score from the training set. The score does not deviate from a normal distribution. B) A histogram of the scores from the training set.
Figure S2. The survival of all arms of the BELOB trial, used for validation.
Figure S3. In order to test whether the profile identifies a group of responders or non‐responders specifically we performed log‐rank of the responders from the arms treated with BEV (alone or in combination) vs the non‐responders from the CCNU arm. This was not significant suggesting that certain patients should not be treated with bev. Additionally these results indicate that even the favorable group treated with Bev show no more improvement in survival than those treated with CCNU, apart from the long‐term survivors.
Table S1. The clinical features of the patients used to test the specificity of the signature to bevacizumab.
Table S2. The risk score was plotted as a density plot, which highlighted 5 minima of density for the risk scores. These risk scores were assessed for their ability to separate the patients into responder and non‐responder groups. The first column represents which minimum this refers to in Figure 1, numbered from left to right. As minimum 4 showed the most significance, with the most powerful hazard ratio, the risk score at this point (0) was used as a cut‐off to separate the training set into responders and non‐responders. This produced 29 ‘responder’ patients with a risk score below 0, and 21 ‘non‐responders’ with a risk score above 1.
Table S3. Multivariate Cox regression for the microRNA profile and the type of treatment received. These are the results when all covariates were included in one model.
Supplementary data
Supplementary data
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2016.06.004.
Hayes Josie, Thygesen Helene, Gregory Walter, Westhead David R., French Pim J., Van Den Bent Martin J., Lawler Sean E., Short Susan C., (2016), A validated microRNA profile with predictive potential in glioblastoma patients treated with bevacizumab, Molecular Oncology, 10, doi: 10.1016/j.molonc.2016.06.004.
References
- Afonyushkin, T. , Oskolkova, O.V. , Bochkov, V.N. , 2012. Permissive role of miR-663 in induction of VEGF and activation of the ATF4 branch of unfolded protein response in endothelial cells by oxidized phospholipids. Atherosclerosis 225, 50–55. 10.1016/j.atherosclerosis.2012.06.016 [DOI] [PubMed] [Google Scholar]
- Babae, N. , Bourajjaj, M. , Liu, Y. , Van Beijnum, J.R. , Cerisoli, F. , Scaria, P.V. , Verheul, M. , Van Berkel, M.P. , Pieters, E.H.E. , Van Haastert, R.J. , Yousefi, A. , Mastrobattista, E. , Storm, G. , Berezikov, E. , Cuppen, E. , Woodle, M. , Schaapveld, R.Q.J. , Prevost, G.P. , Griffioen, A.W. , Van Noort, P.I. , Schiffelers, R.M. , 2014. Systemic miRNA-7 delivery inhibits tumor angiogenesis and growth in murine xenograft glioblastoma. Oncotarget 5, 6687–6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucay, N. , Shahryari, V. , Majid, S. , Yamamura, S. , Mitsui, Y. , Tabatabai, Z.L. , Greene, K. , Deng, G. , Dahiya, R. , Tanaka, Y. , Saini, S. , 2015. miRNA Expression Analyses in Prostate Cancer Clinical Tissues JoVE; 10.3791/53123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network, 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068. 10.1038/nature07385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinot, O.L. , Wick, W. , Mason, W. , Henriksson, R. , Saran, F. , Nishikawa, R. , Carpentier, A.F. , Hoang-Xuan, K. , Kavan, P. , Cernea, D. , Brandes, A.A. , Hilton, M. , Abrey, L. , Cloughesy, T. , 2014. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N. Engl. J. Med. 370, 709–722. 10.1056/NEJMoa1308345 [DOI] [PubMed] [Google Scholar]
- Colman, H. , Zhang, L. , Sulman, E.P. , McDonald, J.M. , Shooshtari, N.L. , Rivera, A. , Popoff, S. , Nutt, C.L. , Louis, D.N. , Cairncross, J.G. , Gilbert, M.R. , Phillips, H.S. , Mehta, M.P. , Chakravarti, A. , Pelloski, C.E. , Bhat, K. , Feuerstein, B.G. , Jenkins, R.B. , Aldape, K. , 2010. A multigene predictor of outcome in glioblastoma. Neuro-oncology 12, 49–57. 10.1093/neuonc/nop007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, A. , Afonso, J. , Osorio, C. , Gomes, A.L. , Caiado, F. , Valente, J. , Aguiar, S.I. , Pinto, F. , Ramirez, M. , Dias, S. , 2013. miR-363-5p regulates endothelial cell properties and their communication with hematopoietic precursor cells. J. Hematol. Oncol. 6, 87 10.1186/1756-8722-6-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano, A.L. , Labussiere, M. , Lombardi, G. , Eoli, M. , Bianchessi, D. , Pasqualetti, F. , Farina, P. , Cuzzubbo, S. , Gállego Pérez-Larraya, J. , Boisselier, B. , Ducray, F. , Cheneau, C. , Moglia, A. , Finocchiaro, G. , Marie, Y. , Rahimian, A. , Hoang-Xuan, K. , Delattre, J.Y. , Mokhtari, K. , Sanson, M. , 2014. VEGFA SNP rs2010963 is associated with vascular toxicity in recurrent glioblastomas and longer response to bevacizumab. J. Neurooncol 10.1007/s11060-014-1677-x [DOI] [PubMed] [Google Scholar]
- Eraslan, L.E. , van den Bent, M. , Kros, J. , 2014. Ge-10 identification of patients with recurrent gbm that benefit from bevacizumab. Neuro-Oncology 10.1093/neuonc/nou256 [Google Scholar]
- Erdem-Eraslan, L. , van den Bent, M.J. , Hoogstrate, Y. , Naz-Khan, H. , Stubbs, A. , van der Spek, P. , Bottcher, R. , Gao, Y. , de Wit, M. , Taal, W. , Oosterkamp, H.M. , Walenkamp, A. , Beerepoot, L.V. , Hanse, M.C.J. , Buter, J. , Honkoop, A.H. , van der Holt, B. , Vernhout, R.M. , Smitt, P.A.E.S. , Kros, J.M. , French, P.J. , 2016. Identification of patients with recurrent glioblastoma who may benefit from combined bevacizumab and CCNU therapy: a report from the BELOB trial. Cancer Res. 76, 525–534. 10.1158/0008-5472.CAN-15-0776 [DOI] [PubMed] [Google Scholar]
- Field, K.M. , Jordan, J.T. , Wen, P.Y. , Rosenthal, M.A. , Reardon, D.A. , 2014. Bevacizumab and glioblastoma: scientific review, newly reported updates, and ongoing controversies. Cancer 10.1002/cncr.28935 n/a–n/a [DOI] [PubMed] [Google Scholar]
- Gilbert, M.R. , Dignam, J.J. , Armstrong, T.S. , Wefel, J.S. , Blumenthal, D.T. , Vogelbaum, M.A. , Colman, H. , Chakravarti, A. , Pugh, S. , Won, M. , Jeraj, R. , Brown, P.D. , Jaeckle, K.A. , Schiff, D. , Stieber, V.W. , Brachman, D.G. , Werner-Wasik, M. , Tremont-Lukats, I.W. , Sulman, E.P. , Aldape, K.D. , Curran, W.J.J. , Mehta, M.P. , 2014. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N. Engl. J. Med. 370, 699–708. 10.1056/NEJMoa1308573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glade-Bender, J. , Kandel, J.J. , Yamashiro, D.J. , 2003. VEGF blocking therapy in the treatment of cancer. Expert Opin. Biol. Ther. 3, 263–276. 10.1517/14712598.3.2.263 [DOI] [PubMed] [Google Scholar]
- Goeman, J.J. , 2010. L1 Penalized Estimation in the Cox Proportional Hazards Model. [DOI] [PubMed] [Google Scholar]
- Gravendeel, L.A.M. , de Rooi, J.J. , Eilers, P.H.C. , van den Bent, M.J. , Smitt, P.A.E.S. , French, P.J. , 2011. Gene expression profiles of gliomas in formalin-fixed paraffin-embedded material. Br. J. Cancer 106, 538–545. 10.1038/bjc.2011.547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, J.S. , Taylor, J. , Valentine, H.R. , Irlam, J.J. , Eustace, A. , Hoskin, P.J. , Miller, C.J. , West, C.M.L. , 2012. Enhanced stability of microRNA expression facilitates classification of FFPE tumour samples exhibiting near total mRNA degradation. Br. J. Cancer 107, 684–694. 10.1038/bjc.2012.294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, J. , Thygesen, H. , Tumilson, C. , Droop, A. , Boissinot, M. , Hughes, T.A. , Westhead, D. , Alder, J.E. , Shaw, L. , Short, S.C. , Lawler, S.E. , 2014. ScienceDirect. Mol. Oncol. 1–11, 10.1016/j.molonc.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam, J.S. , Plyler, J.R. , Bansal, H. , Prajapati, S. , Bansal, S. , Rebeles, J. , Chen, H.-I.H. , Chang, Y.-F. , Panneerdoss, S. , Zoghi, B. , Buddavarapu, K.C. , Broaddus, R. , Hornsby, P. , Tomlinson, G. , Dome, J. , Vadlamudi, R.K. , Pertsemlidis, A. , Chen, Y. , Rao, M.K. , 2012. Genomic loss of tumor suppressor miRNA-204 promotes cancer cell migration and invasion by activating AKT/mTOR/Rac1 signaling and actin reorganization. PLoS One 7, e52397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kather, J.N. , Friedrich, J. , Woik, N. , Sticht, C. , Gretz, N. , Hammes, H.-P. , Kroll, J. , 2014. Angiopoietin-1 is regulated by miR-204 and contributes to corneal neovascularization in KLEIP-deficient mice. Invest Ophthalmol. Vis. Sci. 55, 4295–4303. 10.1167/iovs.13-13619 [DOI] [PubMed] [Google Scholar]
- Kuo, T.-C. , Tan, C.-T. , Chang, Y.-W. , Hong, C.-C. , Lee, W.-J. , Chen, M.-W. , Jeng, Y.-M. , Chiou, J. , Yu, P. , Chen, P.-S. , Wang, M.-Y. , Hsiao, M. , Su, J.-L. , Kuo, M.-L. , 2013. Angiopoietin-like protein 1 suppresses SLUG to inhibit cancer cell motility. J. Clin. Invest 123, 1082–1095. 10.1172/JCI64044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, B.P. , Shih, I.-H. , Jones-Rhoades, M.W. , Bartel, D.P. , Burge, C.B. , 2003. Prediction of mammalian microRNA targets. Cell 115, 787–798. [DOI] [PubMed] [Google Scholar]
- Liao, Y. , Smyth, G.K. , Shi, W. , 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Nakamura, K. , Martin, K.C. , Jackson, J.K. , Beppu, K. , Woo, C.-W. , Thiele, C.J. , 2006. Brain-derived neurotrophic factor activation of TrkB induces vascular endothelial growth factor expression via hypoxia-inducible factor-1alpha in neuroblastoma cells. Cancer Res. 66, 4249–4255. 10.1158/0008-5472.CAN-05-2789 [DOI] [PubMed] [Google Scholar]
- Omuro, A. , Beal, K. , Gutin, P. , Karimi, S. , Correa, D.D. , Kaley, T.J. , DeAngelis, L.M. , Chan, T.A. , Gavrilovic, I.T. , Nolan, C. , Hormigo, A. , Lassman, A.B. , Mellinghoff, I. , Grommes, C. , Reiner, A.S. , Panageas, K.S. , Baser, R.E. , Tabar, V. , Pentsova, E. , Sanchez, J. , Barradas-Panchal, R. , Zhang, J. , Faivre, G. , Brennan, C.W. , Abrey, L.E. , Huse, J.T. , 2014. Phase II study of bevacizumab, temozolomide, and hypofractionated stereotactic radiotherapy for newly diagnosed glioblastoma. Clin. Cancer Res. 20, 5023–5031. 10.1158/1078-0432.CCR-14-0822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevopoulou, M.D. , Georgakilas, G. , Kostoulas, N. , Vlachos, I.S. , Vergoulis, T. , Reczko, M. , Filippidis, C. , Dalamagas, T. , Hatzigeorgiou, A.G. , 2013. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 41, W169–W173. 10.1093/nar/gkt393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao, Y. , Liang, J. , Holmes, L. , Henry, V. , Sulman, E. , de Groot, J.F. , 2013. Acquired resistance to anti-VEGF therapy in glioblastoma is associated with a mesenchymal transition. Clin. Cancer Res. 19, 4392–4403. 10.1158/1078-0432.CCR-12-1557 [DOI] [PubMed] [Google Scholar]
- Piao, Y. , Liang, J. , Holmes, L. , Zurita, A.J. , Henry, V. , Heymach, J.V. , de Groot, J.F. , 2012. Glioblastoma resistance to anti-VEGF therapy is associated with myeloid cell infiltration, stem cell accumulation, and a mesenchymal phenotype. Neuro-oncology 14, 1379–1392. 10.1093/neuonc/nos158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prados, M.D. , Byron, S.A. , Tran, N.L. , Phillips, J.J. , Molinaro, A.M. , Ligon, K.L. , Wen, P.Y. , Kuhn, J.G. , Mellinghoff, I.K. , de Groot, J.F. , Colman, H. , Cloughesy, T.F. , Chang, S.M. , Ryken, T.C. , Tembe, W.D. , Kiefer, J.A. , Berens, M.E. , Craig, D.W. , Carpten, J.D. , Trent, J.M. , 2015. Toward precision medicine in glioblastoma: the promise and the challenges. Neuro-oncology 10.1093/neuonc/nov031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, J. , Roberts, H. , Burton, J. , Pan, J. , States, V. , Rai, S.N. , Galandiuk, S. , 2015. Assay reproducibility in clinical studies of plasma miRNA. PLoS One 10, e0121948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann, T. , Bourgon, R. , Garcia, J. , Li, C. , Cloughesy, T. , Chinot, O.L. , Wick, W. , Nishikawa, R. , Mason, W. , Henriksson, R. , Saran, F. , Lai, A. , Moore, N. , Kharbanda, S. , Peale, F. , Hegde, P. , Abrey, L.E. , Phillips, H.S. , Bais, C. , 2015. Patients with proneural glioblastoma may derive overall survival benefit from the addition of bevacizumab to first-line radiotherapy and temozolomide: retrospective analysis of the AVAglio trial. J. Clin. Oncol. 10.1200/JCO.2015.61.5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Z. , Chen, Q. , Li, C. , Wang, L. , Qian, X. , Jiang, C. , Liu, X. , Wang, X. , Li, H. , Kang, C. , Jiang, T. , Liu, L.-Z. , You, Y. , Liu, N. , Jiang, B.-H. , 2014. MiR-124 governs glioma growth and angiogenesis and enhances chemosensitivity by targeting R-Ras and N-Ras. Neuro-oncology 16, 1341–1353. 10.1093/neuonc/nou084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taal, W. , Oosterkamp, H.M. , Walenkamp, A.M.E. , Dubbink, H.J. , Beerepoot, L.V. , Hanse, M.C.J. , Buter, J. , Honkoop, A.H. , Boerman, D. , de Vos, F.Y.F. , Dinjens, W.N.M. , Enting, R.H. , Taphoorn, M.J.B. , van den Berkmortel, F.W.P.J. , Jansen, R.L.H. , Brandsma, D. , Bromberg, J.E.C. , van Heuvel, I. , Vernhout, R.M. , van der Holt, B. , Van den Bent, M.J. , 2014. Single-agent bevacizumab or lomustine versus a combination of bevacizumab plus lomustine in patients with recurrent glioblastoma (BELOB trial): a randomised controlled phase 2 trial. Lancet Oncol. 15, 943–953. 10.1016/S1470-2045(14)70314-6 [DOI] [PubMed] [Google Scholar]
- Tabouret, E. , Boudouresque, F. , Barrie, M. , Matta, M. , Boucard, C. , Loundou, A. , Carpentier, A. , Sanson, M. , Metellus, P. , Figarella-Branger, D. , Ouafik, L. , Chinot, O. , 2014. Association of matrix metalloproteinase 2 plasma level with response and survival in patients treated with bevacizumab for recurrent high-grade glioma. Neuro-oncology 16, 392–399. 10.1093/neuonc/not226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani, R. , 1996. Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. Ser. B-Methodol. 58, 267–288. 10.1021/np300902g [Google Scholar]
- Trapnell, C. , Pachter, L. , Salzberg, S.L. , 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos, I.S. , Kostoulas, N. , Vergoulis, T. , Georgakilas, G. , Reczko, M. , Maragkakis, M. , Paraskevopoulou, M.D. , Prionidis, K. , Dalamagas, T. , Hatzigeorgiou, A.G. , 2012. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 40, W498–W504. 10.1093/nar/gks494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick, W. , Brandes, A.A. , Gorlia, T. , Bendszus, M. , Sahm, F. , Taal, W. , Taphoorn, M. , Domont, J. , Idbaih, A. , Campone, M. , Clement, P.M. , Stupp, R. , Fabbro, M. , Dubois, F. , Bais, C. , Musmeci, D. , Platten, M. , Weller, M. , Golfinopoulos, V. , van den Bent, M. , 2015. LB-05 Phase III trial exploring the combination of bevacizumab and lomustine in patients with first recurrence of glioblastoma. THE EORTC 26101 TRIAL. Neuro-oncology 17, 10.1093/neuonc/nov306 v1.5–v1 [Google Scholar]
- Zhang, Q. , Kandic, I. , Kutryk, M.J. , 2011. Dysregulation of angiogenesis-related microRNAs in endothelial progenitor cells from patients with coronary artery disease. Biochem. Biophys. Res. Commun. 405, 42–46. 10.1016/j.bbrc.2010.12.119 [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Zhu, C.-D. , Yan, B. , Zhao, J.-L. , Wang, Z.-H. , 2014. miRNA-directed regulation of VEGF in tilapia under hypoxia condition. Biochem. Biophys. Res. Commun. 454, 183–188. 10.1016/j.bbrc.2014.10.068 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Figure S1. A) The distribution of the score from the training set. The score does not deviate from a normal distribution. B) A histogram of the scores from the training set.
Figure S2. The survival of all arms of the BELOB trial, used for validation.
Figure S3. In order to test whether the profile identifies a group of responders or non‐responders specifically we performed log‐rank of the responders from the arms treated with BEV (alone or in combination) vs the non‐responders from the CCNU arm. This was not significant suggesting that certain patients should not be treated with bev. Additionally these results indicate that even the favorable group treated with Bev show no more improvement in survival than those treated with CCNU, apart from the long‐term survivors.
Table S1. The clinical features of the patients used to test the specificity of the signature to bevacizumab.
Table S2. The risk score was plotted as a density plot, which highlighted 5 minima of density for the risk scores. These risk scores were assessed for their ability to separate the patients into responder and non‐responder groups. The first column represents which minimum this refers to in Figure 1, numbered from left to right. As minimum 4 showed the most significance, with the most powerful hazard ratio, the risk score at this point (0) was used as a cut‐off to separate the training set into responders and non‐responders. This produced 29 ‘responder’ patients with a risk score below 0, and 21 ‘non‐responders’ with a risk score above 1.
Table S3. Multivariate Cox regression for the microRNA profile and the type of treatment received. These are the results when all covariates were included in one model.
Supplementary data
Supplementary data


