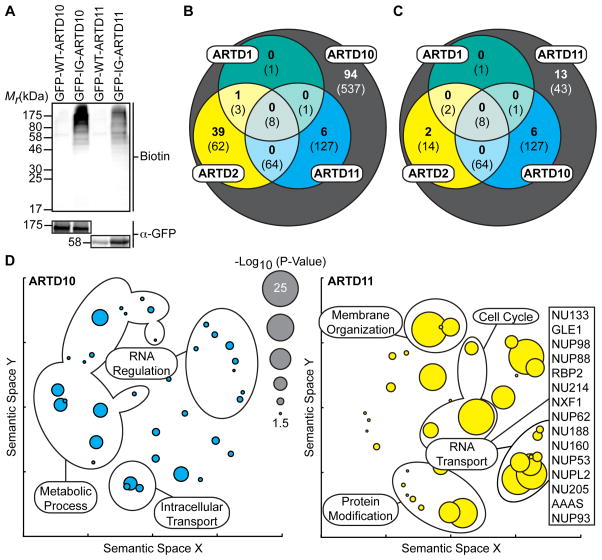
Figure 3. IG-ARTD10 and IG-ARTD11 MARylate Separate, and Family-Member Specific, Protein Targets.
(A) Lysate labeling by IG-ARTD10 or IG-ARTD11 in the presence of 5-Bn-6-a-NAD+. HEK 293T cells were transfected with either WT- or IG-ARTD10 or -ARTD11 and the resulting lysate was incubated for 2 hours in the presence of 5-Bn-6-a-NAD+. MARylation of direct protein targets was observed using streptavidin-HRP (Biotin). The faint bands in the WT-ARTD lanes correspond to endogenous biotinylated proteins. Expression of each ARTD was confirmed via immunoblot detection of GFP. Shown is a representative image from duplicate experiments.
(B) Venn diagram comparing the total IG-ARTD10 target pool with both the current IG-ARTD11 and the previously identified KA-ARTD1 and KA-ARTD2 (Carter-O’Connell et al., 2014) target pools. The protein counts in bold represent the protein targets identified in both LC-MS/MS IG-ARTD10 replicates while the counts in parentheses represent targets identified in at least one replicate. IG-ARTD10 specific targets are shown in the gray circle.
(C) IG-ARTD11 LC-MS/MS targets treated as in (B).
(D) Circle plots depicting enriched GO terms attached to the IG-ARTD10 (left, cyan) or IG-ARTD11 (right, yellow) specific LC-MS/MS identified targets in either replicate. GO term enrichment was performed using the PANTHER toolkit. Significantly enriched GO terms (p < 0.05) were condensed using Revigo and similar terms were plotted based on semantic similarity. Select groups of terms are indicated. Circle radii are scaled proportionally to the −log10(p-value). The IG-ARTD11 specific proteins associated with RNA transport are listed.