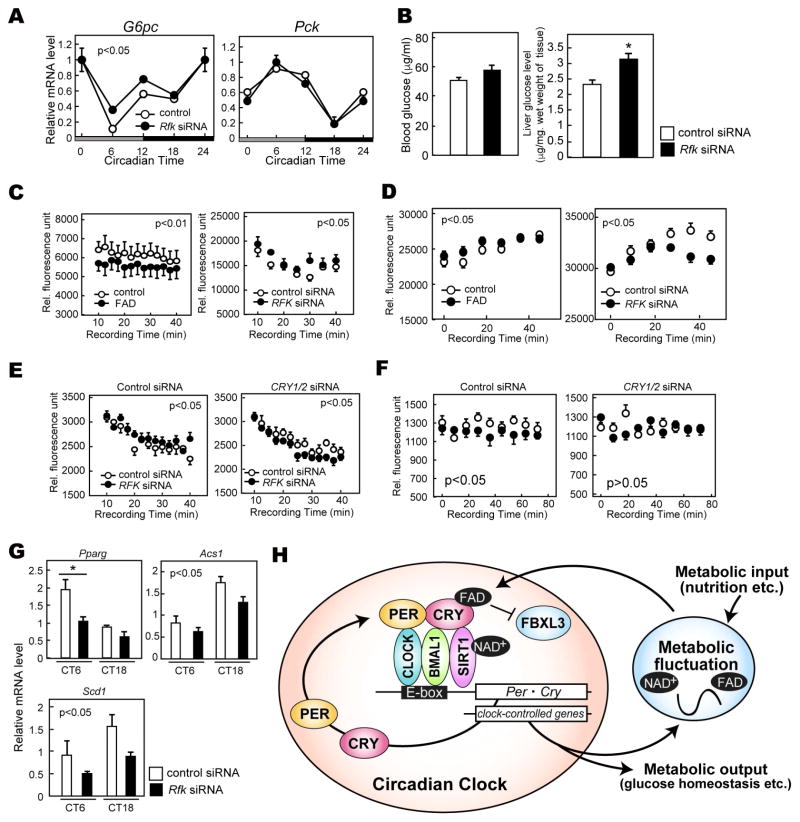
Figure 5. Rfk knockdown results in altered expression of genes in metabolic pathways.
(A) mRNA levels of G6pc and Pck in mouse liver. mRNA levels were normalized to Tbp. Data are shown as means ± SEM (n=3, *: p<0.05 by two-way ANOVA). (B) Glucose levels in blood serum (left) and liver (right) at CT12. Data are shown as means ± SEM (n=3, *: p<0.05 by Student’s t-test) (C) Oxygen consumption rate assay in HepG2 cells. Cells were transfected with siRNA of Rfk or treated with 100 μM FAD for 24 hours. Data are shown as means ± SEM (n=6). Statistical significance was determined by two-way ANOVA. (D) Extracellular acidification rate assay in HepG2 cells. Cells were transfected with Rfk siRNA or treated with 100 μM FAD for 24 hours before recording fluorescence of a glycolysis probe. Data are shown as means ± SEM (n=5 for FAD treatment, n=9 for Rfk knockdown). Statistical significance was determined by two-way ANOVA. (E) Oxygen consumption rate assay in HepG2 cells transfected with siRNA of Rfk and/or CRY1/2. Data are shown as means ± SEM (n=9) (F) Extracellular acidification rate assay in HepG2 cells transfected with siRNA of Rfk and/or CRY1/2. Data are shown as means ± SEM (n=9) (G) mRNA levels of indicated metabolism-relevant genes in mouse liver. mRNA levels were normalized to Tbp. Data are shown as means ± SEM (n=3, *: p<0.05 by two-way ANOVA). (H) Proposed model of CRY regulation by FAD. The classical clock generates gene expression rhythms including Rfk and Nampt, which rhythmically synthesize FAD and NAD+. FAD binding to CRY prevents FBXL3-mediated CRY degradation, leading to stabilization. Time-dependent FAD synthesis modulates CRY expression rhythms and amplitudes, which regulate the gene expression rhythms of important genes for metabolism.