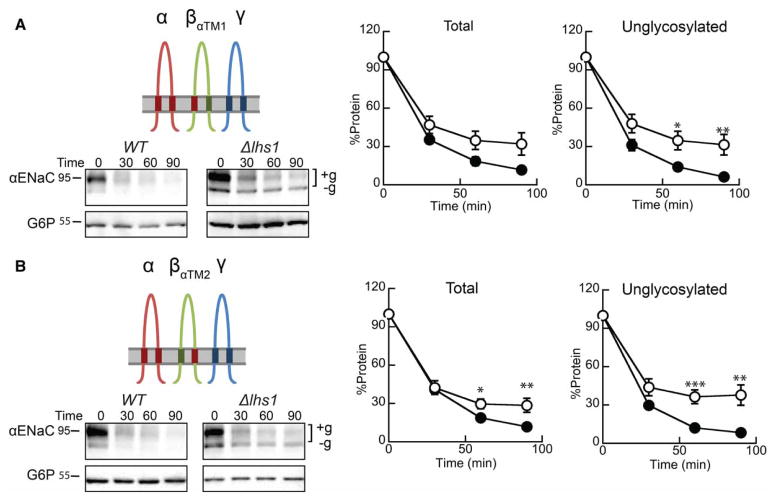
Figure 7. Inserting the α-subunit TMD1 or TMD2 into βENaC restores Lhs1-dependent degradation of αENaC.
Cycloheximide chase reactions were performed as described in the Experimental Procedures section in WT (filled circles) and Δlhs1 (open circles) yeast strains transformed with plasmids designed to express αENaC-HA, γENaC-V5 and (A) βENaCαTM1-13myc or (B) βENaCαTM2-13myc. Lysates were prepared and resolved proteins were immunoblotted with anti-HA antisera (αENaC) and with anti-G6P as a loading control. The expression of myc-tagged and γENaC proteins were confirmed by western blot (data not shown). Glycosylated (+g) and unglycosylated (−g) species are indicated. Data represent the means of 8–11 experiments, ±SEM. *P < 0.05, **P < 0.01, ***P < 0.001.